Q9FKZ0
Gene name |
At5g66910 (MUD21.17) |
Protein name |
Probable disease resistance protein At5g66910 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G66910 |
EC number |
|
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
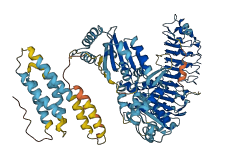
Autoinhibited structure

Activated structure

1 structures for Q9FKZ0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9FKZ0-F1 | Predicted | AlphaFoldDB |
87 variants for Q9FKZ0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH03486651 | 6 | W>L | No | 1000Genomes | |
| ENSVATH03486650 | 25 | V>I | No | 1000Genomes | |
| ENSVATH03486649 | 41 | N>H | No | 1000Genomes | |
| ENSVATH14652194 | 46 | T>M | No | 1000Genomes | |
| ENSVATH00756572 | 50 | L>V | No | 1000Genomes | |
| ENSVATH07492530 | 55 | K>T | No | 1000Genomes | |
| tmp_5_26720935_G_A | 67 | Q>* | No | 1000Genomes | |
| ENSVATH00756571 | 75 | K>E | No | 1000Genomes | |
| tmp_5_26720910_T_A | 75 | K>I | No | 1000Genomes | |
| tmp_5_26720905_G_A | 77 | L>F | No | 1000Genomes | |
| ENSVATH12960552 | 78 | L>V | No | 1000Genomes | |
| ENSVATH00756570 | 91 | I>M | No | 1000Genomes | |
| ENSVATH12960551 | 91 | I>R | No | 1000Genomes | |
| ENSVATH03486646 | 93 | L>M | No | 1000Genomes | |
| ENSVATH07492529 | 100 | K>Q | No | 1000Genomes | |
| ENSVATH12960550 | 107 | K>M | No | 1000Genomes | |
| ENSVATH03486645 | 118 | L>I | No | 1000Genomes | |
| tmp_5_26720760_A_G | 125 | L>S | No | 1000Genomes | |
| ENSVATH07492527 | 133 | A>V | No | 1000Genomes | |
| ENSVATH03486644 | 134 | I>F | No | 1000Genomes | |
| ENSVATH03486644 | 134 | I>L | No | 1000Genomes | |
| ENSVATH03486643 | 135 | L>I | No | 1000Genomes | |
| ENSVATH03486642 | 138 | Y>D | No | 1000Genomes | |
| ENSVATH03486642 | 138 | Y>H | No | 1000Genomes | |
| ENSVATH03486641 | 139 | F>C | No | 1000Genomes | |
| ENSVATH03486640 | 141 | N>I | No | 1000Genomes | |
| ENSVATH07492526 | 142 | I>T | No | 1000Genomes | |
| ENSVATH03486639 | 143 | N>I | No | 1000Genomes | |
| ENSVATH00756569 | 144 | K>E | No | 1000Genomes | |
| ENSVATH00756568 | 148 | R>L | No | 1000Genomes | |
| ENSVATH07492525 | 151 | G>V | No | 1000Genomes | |
| ENSVATH00756567 | 156 | P>A | No | 1000Genomes | |
| ENSVATH12960548 | 167 | L>V | No | 1000Genomes | |
| ENSVATH00756566 | 168 | D>N | No | 1000Genomes | |
| ENSVATH00756565 | 216 | E>K | No | 1000Genomes | |
| ENSVATH00756563 | 229 | N>S | No | 1000Genomes | |
| tmp_5_26720362_G_C | 230 | T>S | No | 1000Genomes | |
| ENSVATH07492521 | 234 | R>K | No | 1000Genomes | |
| ENSVATH07492520 | 235 | A>V | No | 1000Genomes | |
| ENSVATH07492519 | 254 | D>E | No | 1000Genomes | |
| ENSVATH07492518 | 291 | K>N | No | 1000Genomes | |
| ENSVATH14652192 | 301 | I>N | No | 1000Genomes | |
| tmp_5_26720126_A_C | 309 | F>V | No | 1000Genomes | |
| ENSVATH07492517 | 324 | E>K | No | 1000Genomes | |
| ENSVATH07492516 | 325 | Y>D | No | 1000Genomes | |
| ENSVATH12960546 | 345 | Y>C | No | 1000Genomes | |
| tmp_5_26720012_C_T | 347 | D>N | No | 1000Genomes | |
| tmp_5_26719896_C_T | 355 | R>H | No | 1000Genomes | |
| tmp_5_26719840_C_A | 374 | A>S | No | 1000Genomes | |
| ENSVATH07492508 | 395 | A>P | No | 1000Genomes | |
| ENSVATH00756562 | 396 | N>T | No | 1000Genomes | |
| ENSVATH14652191 | 408 | N>S | No | 1000Genomes | |
| ENSVATH12960534 | 422 | G>S | No | 1000Genomes | |
| ENSVATH07492507 | 427 | D>G | No | 1000Genomes | |
| ENSVATH00756561 | 434 | L>V | No | 1000Genomes | |
| ENSVATH07492506 | 448 | S>T | No | 1000Genomes | |
| tmp_5_26719607_G_C | 451 | N>K | No | 1000Genomes | |
| ENSVATH07492503 | 458 | N>K | No | 1000Genomes | |
| ENSVATH12960533 | 460 | L>I | No | 1000Genomes | |
| ENSVATH07492498 | 475 | K>M | No | 1000Genomes | |
| ENSVATH07492495 | 490 | N>D | No | 1000Genomes | |
| ENSVATH07492494 | 514 | E>A | No | 1000Genomes | |
| ENSVATH07492490 | 545 | K>N | No | 1000Genomes | |
| ENSVATH07492489 | 547 | L>V | No | 1000Genomes | |
| ENSVATH07492487 | 548 | E>K | No | 1000Genomes | |
| tmp_5_26719125_A_G | 554 | V>A | No | 1000Genomes | |
| ENSVATH07492486 | 556 | A>T | No | 1000Genomes | |
| ENSVATH12960521 | 556 | A>V | No | 1000Genomes | |
| ENSVATH07492485 | 574 | E>G | No | 1000Genomes | |
| ENSVATH07492484 | 584 | A>T | No | 1000Genomes | |
| ENSVATH12960517 | 592 | R>S | No | 1000Genomes | |
| tmp_5_26718987_C_G | 600 | S>T | No | 1000Genomes | |
| tmp_5_26718978_G_A | 603 | P>L | No | 1000Genomes | |
| tmp_5_26718979_G_A | 603 | P>S | No | 1000Genomes | |
| ENSVATH07492481 | 624 | Q>L | No | 1000Genomes | |
| tmp_5_26718846_T_C | 647 | D>G | No | 1000Genomes | |
| ENSVATH07492479 | 676 | P>S | No | 1000Genomes | |
| ENSVATH14652187 | 679 | V>I | No | 1000Genomes | |
| tmp_5_26718705_T_G | 694 | Q>P | No | 1000Genomes | |
| ENSVATH12960515 | 710 | M>I | No | 1000Genomes | |
| tmp_5_26718604_T_A | 728 | N>Y | No | 1000Genomes | |
| ENSVATH03486632 | 754 | E>D | No | 1000Genomes | |
| ENSVATH14652185 | 768 | D>N | No | 1000Genomes | |
| ENSVATH03486631 | 777 | E>K | No | 1000Genomes | |
| ENSVATH03486630 | 778 | V>I | No | 1000Genomes | |
| tmp_5_26718391_T_C | 799 | R>G | No | 1000Genomes | |
| tmp_5_26718340_A_C | 816 | F>E | No | 1000Genomes |
No associated diseases with Q9FKZ0
1 regional properties for Q9FKZ0
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | ZipA, C-terminal FtsZ-binding domain | 191 - 320 | IPR007449 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endomembrane system | A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ADP binding | Binding to ADP, adenosine 5'-diphosphate. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ubiquitin binding | Binding to ubiquitin, a protein that when covalently bound to other cellular proteins marks them for proteolytic degradation. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| defense response to fungus | Reactions triggered in response to the presence of a fungus that act to protect the cell or organism. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| F1MHT9 | NLRC4 | NLR family CARD domain-containing protein 4 | Bos taurus (Bovine) | SS |
| Q9NPP4 | NLRC4 | NLR family CARD domain-containing protein 4 | Homo sapiens (Human) | SS |
| Q3UP24 | Nlrc4 | NLR family CARD domain-containing protein 4 | Mus musculus (Mouse) | EV |
| F1M649 | Nlrc4 | NLR family CARD domain-containing protein 4 | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVVVDWLGLG | LGSVAGALVS | EGLKVLISEA | KKVLAFKSVS | NELASTMESL | LPVIKEIESM |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QDGMELQDLK | DTIDKALLLV | EKCSHVEKWN | IILKSKYTRK | VEEINRKMLK | FCQVQLQLLL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| FRNQLKSMPS | MEAILNNYFQ | NINKKLDRLS | GSPAPPLVSK | RCSVPKLDNM | VLVGLDWPLV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ELKKKLLDNS | VVVVSGPPGC | GKTTLVTKLC | DDPEIEGEFK | KIFYSVVSNT | PNFRAIVQNL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LQDNGCGAIT | FDDDSQAETG | LRDLLEELTK | DGRILLVLDD | VWQGSEFLLR | KFQIDLPDYK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ILVTSQFDFT | SLWPTYHLVP | LKYEYARSLL | IQWASPPLHT | SPDEYEDLLQ | KILKRCNGFP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LVIEVVGISL | KGQALYLWKG | QVESWSEGET | ILGNANPTVR | QRLQPSFNVL | KPHLKECFMD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| MGSFLQDQKI | RASLIIDIWM | ELYGRGSSST | NKFMLYLNEL | ASQNLLKLVH | LGTNKREDGF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YNELLVTQHN | ILRELAIFQS | ELEPIMQRKK | LNLEIREDNF | PDECLNQPIN | ARLLSIYTDD |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LFSSKWLEMD | CPNVEALVLN | ISSLDYALPS | FIAEMKKLKV | LTIANHGFYP | ARLSNFSCLS |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SLPNLKRIRF | EKVSVTLLDI | PQLQLGSLKK | LSFFMCSFGE | VFYDTEDIDV | SKALSNLQEI |
| 670 | 680 | 690 | 700 | 710 | 720 |
| DIDYCYDLDE | LPYWIPEVVS | LKTLSITNCN | KLSQLPEAIG | NLSRLEVLRM | CSCMNLSELP |
| 730 | 740 | 750 | 760 | 770 | 780 |
| EATERLSNLR | SLDISHCLGL | RKLPQEIGKL | QKLENISMRK | CSGCELPDSV | RYLENLEVKC |
| 790 | 800 | 810 | |||
| DEVTGLLWER | LMPEMRNLRV | HTEETEHNLK | LLLTF |