Q9FKW6
Gene name |
LFNR1 |
Protein name |
Ferredoxin--NADP reductase, leaf isozyme 1, chloroplastic |
Names |
Leaf FNR 1, AtLFNR1, FNR-1 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G66190 |
EC number |
1.18.1.2: With NAD(+) or NADP(+) as acceptor |
Protein Class |
|
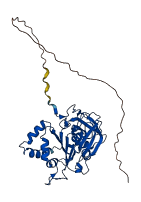
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9FKW6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9FKW6-F1 | Predicted | AlphaFoldDB |
9 variants for Q9FKW6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH03482618 | 20 | L>P | No | 1000Genomes | |
| tmp_5_26452916_G_T | 33 | L>I | No | 1000Genomes | |
| ENSVATH12949480 | 62 | V>L | No | 1000Genomes | |
| tmp_5_26452656_A_G | 84 | Y>H | No | 1000Genomes | |
| ENSVATH14650950 | 165 | V>I | No | 1000Genomes | |
| ENSVATH07488147 | 192 | K>R | No | 1000Genomes | |
| ENSVATH14650948 | 203 | M>I | No | 1000Genomes | |
| ENSVATH07488144 | 289 | E>D | No | 1000Genomes | |
| ENSVATH12949461 | 338 | D>E | No | 1000Genomes |
No associated diseases with Q9FKW6
10 regional properties for Q9FKW6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Oxidoreductase FAD/NAD(P)-binding | 213 - 327 | IPR001433 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 116 - 126 | IPR001709-1 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 139 - 146 | IPR001709-2 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 176 - 185 | IPR001709-3 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 212 - 231 | IPR001709-4 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 242 - 251 | IPR001709-5 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 254 - 265 | IPR001709-6 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 290 - 306 | IPR001709-7 |
| domain | Flavoprotein pyridine nucleotide cytochrome reductase | 314 - 322 | IPR001709-8 |
| domain | FAD-binding domain, ferredoxin reductase-type | 81 - 203 | IPR017927 |
Functions
| Description | ||
|---|---|---|
| EC Number | 1.18.1.2 | With NAD(+) or NADP(+) as acceptor |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| apoplast | The cell membranes and intracellular regions in a plant are connected through plasmodesmata, and plants may be described as having two major compartments: the living symplast and the non-living apoplast. The apoplast is external to the plasma membrane and includes cell walls, intercellular spaces and the lumen of dead structures such as xylem vessels. Water and solutes pass freely through it. |
| chloroplast | A chlorophyll-containing plastid with thylakoids organized into grana and frets, or stroma thylakoids, and embedded in a stroma. |
| chloroplast envelope | The double lipid bilayer enclosing the chloroplast and separating its contents from the rest of the cytoplasm; includes the intermembrane space. |
| chloroplast stroma | The space enclosed by the double membrane of a chloroplast but excluding the thylakoid space. It contains DNA, ribosomes and some temporary products of photosynthesis. |
| chloroplast thylakoid | Sac-like membranous structures (cisternae) in a chloroplast combined into stacks (grana) and present singly in the stroma (stroma thylakoids or frets) as interconnections between grana. An example of this component is found in Arabidopsis thaliana. |
| chloroplast thylakoid membrane | The pigmented membrane of a chloroplast thylakoid. An example of this component is found in Arabidopsis thaliana. |
| chloroplast thylakoid membrane protein complex | A protein complex that is part of a chloroplast thylakoid membrane. |
| plastid | Any member of a family of organelles found in the cytoplasm of plants and some protists, which are membrane-bounded and contain DNA. Plant plastids develop from a common type, the proplastid. |
| thylakoid | A membranous cellular structure that bears the photosynthetic pigments in plants, algae, and cyanobacteria. In cyanobacteria thylakoids are of various shapes and are attached to, or continuous with, the plasma membrane. In eukaryotes they are flattened, membrane-bounded disk-like structures located in the chloroplasts; in the chloroplasts of higher plants the thylakoids form dense stacks called grana. Isolated thylakoid preparations can carry out photosynthetic electron transport and the associated phosphorylation. |
| thylakoid lumen | The volume enclosed by a thylakoid membrane. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity | Enables the directed movement of electrons within the cyclic electron transport pathway of photosynthesis. |
| electron transporter, transferring electrons within the noncyclic electron transport pathway of photosynthesis activity | Enables the directed movement of electrons within the noncyclic electron transport pathway of photosynthesis. |
| ferredoxin-NADP+ reductase activity | Catalysis of the reaction: reduced ferredoxin + NADP+ = oxidized ferredoxin + NADPH + H+. |
| poly(U) RNA binding | Binding to a sequence of uracil residues in an RNA molecule. |
| protein domain specific binding | Binding to a specific domain of a protein. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| photosynthetic electron transport chain | A process, occurring as part of photosynthesis, in which light provides the energy for a series of electron carriers to operate together to transfer electrons and generate a transmembrane electrochemical gradient. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P29473 | NOS3 | Nitric oxide synthase, endothelial | Bos taurus (Bovine) | EV |
| Q9UBK8 | MTRR | Methionine synthase reductase | Homo sapiens (Human) | PR |
| P29474 | NOS3 | Nitric oxide synthase, endothelial | Homo sapiens (Human) | SS |
| P29475 | NOS1 | Nitric oxide synthase, brain | Homo sapiens (Human) | SS |
| P70313 | Nos3 | Nitric oxide synthase 3 | Mus musculus (Mouse) | SS |
| Q8C1A3 | Mtrr | Methionine synthase reductase | Mus musculus (Mouse) | PR |
| Q28969 | NOS3 | Nitric oxide synthase 3 | Sus scrofa (Pig) | SS |
| Q62600 | Nos3 | Nitric oxide synthase 3 | Rattus norvegicus (Rat) | SS |
| Q498R1 | Mtrr | Methionine synthase reductase | Rattus norvegicus (Rat) | PR |
| P29476 | Nos1 | Nitric oxide synthase, brain | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAAISAAVS | LPSSKSSSLL | TKISSVSPQR | IFLKKSTVCY | RRVVSVKAQV | TTDTTEAPPV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KVVKESKKQE | EGIVVNKFKP | KNPYTGRCLL | NTKITGDDAP | GETWHIVFTT | EGEVPYREGQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SIGVIPEGID | KNGKPHKLRL | YSIASSAIGD | FGDSKTVSLC | VKRLVYTNDG | GEIVKGVCSN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FLCDLKPGDE | AKITGPVGKE | MLMPKDPNAT | IIMLGTGTGI | APFRSFLWKM | FFEEHEDYKF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NGLAWLFLGV | PTSSSLLYKE | EFEKMKEKNP | DNFRLDFAVS | REQTNEKGEK | MYIQTRMAEY |
| 310 | 320 | 330 | 340 | 350 | |
| AEELWELLKK | DNTFVYMCGL | KGMEKGIDDI | MVSLAAKDGI | DWLEYKKQLK | RSEQWNVEVY |