Q9FK75
Gene name |
At5g45670 (MRA19.6) |
Protein name |
GDSL esterase/lipase At5g45670 |
Names |
Extracellular lipase At5g45670 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G45670 |
EC number |
|
Protein Class |
|
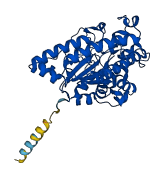
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9FK75
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9FK75-F1 | Predicted | AlphaFoldDB |
22 variants for Q9FK75
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_5_18528606_A_G | 3 | R>G | No | 1000Genomes | |
| ENSVATH00712467 | 5 | S>C | No | 1000Genomes | |
| ENSVATH03365747 | 6 | L>M | No | 1000Genomes | |
| ENSVATH03365747 | 6 | L>V | No | 1000Genomes | |
| ENSVATH07347233 | 7 | M>L | No | 1000Genomes | |
| ENSVATH03365748 | 8 | I>M | No | 1000Genomes | |
| tmp_5_18528633_A_C | 12 | M>L | No | 1000Genomes | |
| ENSVATH00712468 | 15 | V>M | No | 1000Genomes | |
| ENSVATH07347235 | 17 | M>I | No | 1000Genomes | |
| ENSVATH12560518 | 90 | T>I | No | 1000Genomes | |
| ENSVATH14596085 | 192 | D>E | No | 1000Genomes | |
| tmp_5_18529412_G_A | 192 | D>N | No | 1000Genomes | |
| tmp_5_18529424_G_T,A | 196 | A>S | No | 1000Genomes | |
| tmp_5_18529424_G_T,A | 196 | A>T | No | 1000Genomes | |
| tmp_5_18529612_A_T | 208 | N>Y | No | 1000Genomes | |
| ENSVATH14596086 | 214 | A>V | No | 1000Genomes | |
| tmp_5_18529732_A_G | 248 | I>V | No | 1000Genomes | |
| ENSVATH07347262 | 252 | K>N | No | 1000Genomes | |
| tmp_5_18529774_C_A | 262 | Q>K | No | 1000Genomes | |
| ENSVATH00712477 | 266 | D>E | No | 1000Genomes | |
| ENSVATH07347264 | 286 | A>S | No | 1000Genomes | |
| ENSVATH12560552 | 313 | Q>E | No | 1000Genomes |
No associated diseases with Q9FK75
No regional properties for Q9FK75
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q9FK75 | |||
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| hydrolase activity, acting on ester bonds | Catalysis of the hydrolysis of any ester bond. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| lipid catabolic process | The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A4FUB7 | GIN1 | Gypsy retrotransposon integrase-like protein 1 | Bos taurus (Bovine) | PR |
| Q6ZNG9 | KRBA2 | KRAB-A domain-containing protein 2 | Homo sapiens (Human) | PR |
| Q9P2P1 | NYNRIN | Protein NYNRIN | Homo sapiens (Human) | PR |
| Q6R2W3 | ZBED9 | SCAN domain-containing protein 3 | Homo sapiens (Human) | PR |
| Q9UN19 | DAPP1 | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3-phosphoinositide | Homo sapiens (Human) | PR |
| Q9QXT1 | Dapp1 | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3-phosphoinositide | Mus musculus (Mouse) | PR |
| Q94KB1 | MLO14 | MLO-like protein 14 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O80580 | MLO15 | MLO-like protein 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SMP1 | XTH11 | Probable xyloglucan endotransglucosylase/hydrolase protein 11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O49621 | MLO1 | MLO-like protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FI00 | MLO11 | MLO-like protein 11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64468 | At2g19050 | GDSL esterase/lipase At2g19050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MARMSLMIMM | IMVAVTMINI | AKSDPIAPCY | FIFGDSLVDN | GNNNQLQSLA | RANYFPYGID |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FAAGPTGRFS | NGLTTVDVIA | QLLGFEDYIT | PYASARGQDI | LRGVNYASAA | AGIRDETGRQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LGGRIAFAGQ | VANHVNTVSQ | VVNILGDQNE | ASNYLSKCIY | SIGLGSNDYL | NNYFMPTFYS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TGNQFSPESY | ADDLVARYTE | QLRVLYTNGA | RKFALIGVGA | IGCSPNELAQ | NSRDGRTCDE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RINSANRIFN | SKLISIVDAF | NQNTPDAKFT | YINAYGIFQD | IITNPARYGF | RVTNAGCCGV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GRNNGQITCL | PGQAPCLNRN | EYVFWDAFHP | GEAANIVIGR | RSFKREAASD | AHPYDIQQLA |
| SL |