Q9ES70
Gene name |
Nek6 |
Protein name |
Serine/threonine-protein kinase Nek6 |
Names |
Never in mitosis A-related kinase 6, NimA-related protein kinase 6 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:59126 |
EC number |
2.7.11.34: Protein-serine/threonine kinases |
Protein Class |
|
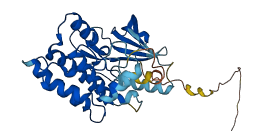
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
31-41 (N-terminal extension of kinase domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
189-212 (Activation loop from InterPro)
Target domain |
45-310 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9ES70
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9ES70-F1 | Predicted | AlphaFoldDB |
No variants for Q9ES70
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q9ES70 | |||||
No associated diseases with Q9ES70
6 regional properties for Q9ES70
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| binding_site | IQ motif, EF-hand binding site | 781 - 803 | IPR000048-1 |
| binding_site | IQ motif, EF-hand binding site | 807 - 829 | IPR000048-2 |
| domain | Myosin head, motor domain | 80 - 780 | IPR001609 |
| domain | Myosin tail | 847 - 1924 | IPR002928 |
| domain | Myosin, N-terminal, SH3-like | 33 - 82 | IPR004009 |
| domain | Class II myosin, Myh3, motor domain | 100 - 767 | IPR036000 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.34 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| centriolar satellite | A small (70-100 nm) cytoplasmic granule that contains a number of centrosomal proteins; centriolar satellites traffic toward microtubule minus ends and are enriched near the centrosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nuclear speck | A discrete extra-nucleolar subnuclear domain, 20-50 in number, in which splicing factors are seen to be localized by immunofluorescence microscopy. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| spindle pole | Either of the ends of a spindle, where spindle microtubules are organized; usually contains a microtubule organizing center and accessory molecules, spindle microtubules and astral microtubules. |
10 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| kinesin binding | Interacting selectively and non-covalently and stoichiometrically with kinesin, a member of a superfamily of microtubule-based motor proteins that perform force-generating tasks such as organelle transport and chromosome segregation. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| transcription corepressor binding | Binding to a transcription corepressor, a protein involved in negative regulation of transcription via protein-protein interactions with transcription factors and other proteins that negatively regulate transcription. Transcription corepressors do not bind DNA directly, but rather mediate protein-protein interactions between repressing transcription factors and the basal transcription machinery. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| chromosome segregation | The process in which genetic material, in the form of chromosomes, is organized into specific structures and then physically separated and apportioned to two or more sets. In eukaryotes, chromosome segregation begins with the condensation of chromosomes, includes chromosome separation, and ends when chromosomes have completed movement to the spindle poles. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of mitotic metaphase/anaphase transition | Any process that modulates the frequency, rate or extent of the cell cycle process in which a cell progresses from metaphase to anaphase during mitosis, triggered by the activation of the anaphase promoting complex by Cdc20/Sleepy homolog which results in the degradation of Securin. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8TDX7 | NEK7 | Serine/threonine-protein kinase Nek7 | Homo sapiens (Human) | EV |
| Q9HC98 | NEK6 | Serine/threonine-protein kinase Nek6 | Homo sapiens (Human) | EV |
| Q00526 | CDK3 | Cyclin-dependent kinase 3 | Homo sapiens (Human) | PR |
| Q9ES74 | Nek7 | Serine/threonine-protein kinase Nek7 | Mus musculus (Mouse) | SS |
| Q61084 | Map3k3 | Mitogen-activated protein kinase kinase kinase 3 | Mus musculus (Mouse) | PR |
| Q61083 | Map3k2 | Mitogen-activated protein kinase kinase kinase 2 | Mus musculus (Mouse) | PR |
| A2BD05 | NEK6 | Serine/threonine-protein kinase Nek6 | Sus scrofa (Pig) | SS |
| D3ZBE5 | Nek7 | Serine/threonine-protein kinase Nek7 | Rattus norvegicus (Rat) | SS |
| P59895 | Nek6 | Serine/threonine-protein kinase Nek6 | Rattus norvegicus (Rat) | SS |
| G5EFM9 | nekl-3 | Serine/threonine-protein kinase nekl-3 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAGQPSHMPH | GGSPNHLCHA | LGPAPPPDPQ | RLPNTLSFRC | SLADFQIEKK | IGRGQFSEVY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KATCLLDRKT | VALKKVQIFE | MMDAKARQDC | VKEIGLLKQL | NHPNIIKYLD | SFIEDNELNI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VLELADAGDL | SQMIKYFKKQ | KRLIPERTVW | KYFVQLCSAV | EHMHSRRVMH | RDIKPANVFI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TATGIVKLGD | LGLGRFFSSE | TTAAHSLVGT | PYYMSPERIH | ENGYNFKSDI | WSLGCLLYEM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AALQSPFYGD | KMNLFSLCQK | IEQCDYPPLP | GEHYSEKLRE | LVSMCIYPDP | DHRPDIVYVH |
| 310 | |||||
| QVARQMHVWT | SST |