Q9ER42
Gene name |
Barx1 |
Protein name |
Homeobox protein BarH-like 1 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:12022 |
EC number |
|
Protein Class |
|
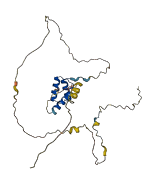
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9ER42
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9ER42-F1 | Predicted | AlphaFoldDB |
6 variants for Q9ER42
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs238928568 | 137 | A>T | No | EVA | |
| rs3389284154 | 216 | R>Q | No | EVA | |
| rs1133699716 | 241 | A>G | No | EVA | |
| rs3404257302 | 246 | E>* | No | EVA | |
| rs3404157257 | 246 | E>A | No | EVA | |
| rs3403717207 | 248 | S>G | No | EVA |
No associated diseases with Q9ER42
5 regional properties for Q9ER42
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Helix-turn-helix motif | 171 - 196 | IPR000047 |
| domain | Homeobox domain | 140 - 204 | IPR001356 |
| conserved_site | Homeobox, conserved site | 175 - 198 | IPR017970 |
| domain | Homeobox domain, metazoa | 164 - 175 | IPR020479-1 |
| domain | Homeobox domain, metazoa | 179 - 198 | IPR020479-2 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
11 GO annotations of biological process
| Name | Definition |
|---|---|
| anatomical structure development | The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome. |
| animal organ development | Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions. |
| anterior/posterior pattern specification | The regionalization process in which specific areas of cell differentiation are determined along the anterior-posterior axis. The anterior-posterior axis is defined by a line that runs from the head or mouth of an organism to the tail or opposite end of the organism. |
| cell-cell signaling | Any process that mediates the transfer of information from one cell to another. This process includes signal transduction in the receiving cell and, where applicable, release of a ligand and any processes that actively facilitate its transport and presentation to the receiving cell. Examples include signaling via soluble ligands, via cell adhesion molecules and via gap junctions. |
| digestive system development | The process whose specific outcome is the progression of the digestive system over time, from its formation to the mature structure. The digestive system is the entire structure in which digestion takes place. Digestion is all of the physical, chemical, and biochemical processes carried out by multicellular organisms to break down ingested nutrients into components that may be easily absorbed and directed into metabolism. |
| endothelial cell differentiation | The process in which a mesodermal, bone marrow or neural crest cell acquires specialized features of an endothelial cell, a thin flattened cell. A layer of such cells lines the inside surfaces of body cavities, blood vessels, and lymph vessels, making up the endothelium. |
| negative regulation of Wnt signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of the Wnt signaling pathway. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| spleen development | The process whose specific outcome is the progression of the spleen over time, from its formation to the mature structure. The spleen is a large vascular lymphatic organ composed of white and red pulp, involved both in hemopoietic and immune system functions. |
| tissue development | The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure. |
| Wnt signaling pathway | The series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell and ending with a change in cell state. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9DED6 | BARX1B | Homeobox protein BarH-like 1b | Gallus gallus (Chicken) | PR |
| Q6RFL5 | BSX | Brain-specific homeobox protein homolog | Gallus gallus (Chicken) | PR |
| Q9W6D8 | BARX1 | Homeobox protein BarH-like 1 | Gallus gallus (Chicken) | PR |
| Q9UMQ3 | BARX2 | Homeobox protein BarH-like 2 | Homo sapiens (Human) | PR |
| Q9HBU1 | BARX1 | Homeobox protein BarH-like 1 | Homo sapiens (Human) | PR |
| Q3C1V8 | BSX | Brain-specific homeobox protein homolog | Homo sapiens (Human) | PR |
| Q810B3 | Bsx | Brain-specific homeobox protein homolog | Mus musculus (Mouse) | PR |
| Q6H6S3 | HOX24 | Homeobox-leucine zipper protein HOX24 | Oryza sativa subsp japonica (Rice) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MQRPGEPGAA | RFGPPEGCAD | HRPHRYRSFM | IEEILTEPPG | PKGAAPAAAA | AAAGELLKFG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VQALLAARPF | HSHLAVLKAE | QAAVFKFPLA | PLGCSGLGSA | LLAAGPGMPG | PAGASHLPLE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LQLRGKLEAA | GSGEPGAKAK | KGRRSRTVFT | ELQLMGLEKR | FEKQKYLSTP | DRIDLAESLG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LSQLQVKTWY | QNRRMKWKKI | VLQGGGLESP | TKPKGRPKKN | SIPTSEQLTE | QERAKETEKP |
| 250 | |||||
| AETPGEPSDR | NCED |