Q9DBX3
Gene name |
Susd2 |
Protein name |
Sushi domain-containing protein 2 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:71733 |
EC number |
|
Protein Class |
|
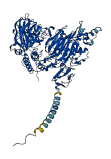
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q9DBX3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2CQW | NMR | - | A | 16-86 | PDB |
| AF-Q9DBX3-F1 | Predicted | AlphaFoldDB |
54 variants for Q9DBX3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389049279 | 17 | P>L | No | EVA | |
| rs3389117624 | 28 | S>T | No | EVA | |
| rs3389113339 | 38 | G>* | No | EVA | |
| rs3389110004 | 39 | L>P | No | EVA | |
| rs3389106174 | 55 | D>E | No | EVA | |
| rs30005620 | 64 | L>S | No | EVA | |
| rs3389102894 | 106 | D>V | No | EVA | |
| rs3389080708 | 114 | S>* | No | EVA | |
| rs3401425060 | 136 | P>T | No | EVA | |
| rs3389085760 | 160 | E>D | No | EVA | |
| rs3389073314 | 166 | Y>F | No | EVA | |
| rs1132036225 | 199 | T>K | No | EVA | |
| rs3389104844 | 243 | V>M | No | EVA | |
| rs3389117632 | 247 | R>G | No | EVA | |
| rs3389080642 | 263 | L>P | No | EVA | |
| rs3389106973 | 277 | D>E | No | EVA | |
| rs242246466 | 280 | A>E | No | EVA | |
| rs3389073247 | 284 | A>G | No | EVA | |
| rs3389117691 | 297 | E>D | No | EVA | |
| rs3401392273 | 305 | T>A | No | EVA | |
| rs252878483 | 305 | T>M | No | EVA | |
| rs3389073272 | 308 | P>S | No | EVA | |
| rs3389115408 | 309 | D>E | No | EVA | |
| rs3389073282 | 326 | T>S | No | EVA | |
| rs3389085752 | 333 | E>D | No | EVA | |
| rs3389049373 | 361 | Q>H | No | EVA | |
| rs3389110013 | 364 | C>R | No | EVA | |
| rs3389110854 | 424 | P>S | No | EVA | |
| rs3389080669 | 428 | K>T | No | EVA | |
| rs3389085815 | 434 | D>Y | No | EVA | |
| rs3389104889 | 446 | A>V | No | EVA | |
| rs3412056149 | 467 | E>* | No | EVA | |
| rs247830180 | 488 | R>K | No | EVA | |
| rs213711685 | 532 | V>A | No | EVA | |
| rs3401425099 | 534 | S>N | No | EVA | |
| rs3401343295 | 536 | T>S | No | EVA | |
| rs3401331212 | 552 | A>T | No | EVA | |
| rs3389073332 | 570 | V>I | No | EVA | |
| rs3389073332 | 570 | V>L | No | EVA | |
| rs3389108112 | 592 | L>F | No | EVA | |
| rs3389110885 | 631 | S>C | No | EVA | |
| rs213504278 | 673 | T>P | No | EVA | |
| rs227244338 | 676 | V>M | No | EVA | |
| rs3389100117 | 685 | F>I | No | EVA | |
| rs3389108047 | 691 | M>I | No | EVA | |
| rs264788252 | 696 | S>P | No | EVA | |
| rs3389073315 | 703 | R>W | No | EVA | |
| rs248365239 | 710 | Q>H | No | EVA | |
| rs3389085802 | 711 | H>Q | No | EVA | |
| rs259342810 | 749 | N>D | No | EVA | |
| rs3389093888 | 783 | V>A | No | EVA | |
| rs3389110003 | 784 | L>I | No | EVA | |
| rs3389049288 | 800 | I>N | No | EVA | |
| rs3389073328 | 815 | M>T | No | EVA |
No associated diseases with Q9DBX3
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular space | That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| negative regulation of cell cycle G1/S phase transition | Any signalling pathway that decreases or inhibits the activity of a cell cycle cyclin-dependent protein kinase to modulate the switch from G1 phase to S phase of the cell cycle. |
| negative regulation of cell division | Any process that stops, prevents, or reduces the frequency, rate or extent of cell division. |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKLALLPWIL | MLLSTIPGPG | FTAGAQGSCS | LRCGAQDGLC | SCHPTCSGLG | TCCEDFLDYC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LEILPSSGSM | MGGKDFVVQH | LKWTDPTDGV | ICRFKESIQT | LGYVDDFYQV | HCISPLLYES |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GYIPFTISMD | NGRSFPHAGT | WLAAHPYKVS | ESEKSQLVNE | THWQYYGTSD | TRGNLNLTWD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TSALPTPAVT | IELWGYEETG | KPYSGNWTSK | WSYLYPLATN | IPNTGFFTFT | PKPASPQYQR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| WKVGALRISS | SRNYPGEKDV | RALWTNDHAL | AWHLGDDFRA | DSVAWARAQC | LAWEALEDQL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PNFLTELPDC | PCTLAQARAD | SGRFFTDYGC | DIEHGSVCTY | HPGAVHCVRS | VQASPRYGSG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QQCCYTAAGT | QLLTSDSTSG | STPDRGHDWG | APPYRTPPRV | PGMSHWLYDV | ISFYYCCLWA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| PECPRYMKRR | PSSDCRNYRP | PRLASAFGDP | HFVTFDGTSF | SFSGNGEYVL | LETTLSDLRV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| QGRAQPGRMP | NGTQARGTGL | TAVAVQEDNS | DVIEVRLAGG | SRVLEVLLNQ | KVLSFTEQNW |
| 550 | 560 | 570 | 580 | 590 | 600 |
| MDLKGMFLSV | AAQDKVSIML | SSGAGLEVGV | QGPFLSVSIL | LPEKFLTHTR | GLLGTLNNNP |
| 610 | 620 | 630 | 640 | 650 | 660 |
| RDDFTLRNGQ | VLPLNASAQQ | VFQFGADWAV | SNTSSLFTYD | SWLLVYQFVY | GPKHNPNFKP |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LFPDETTLSP | SQTEDVARLC | EGDRFCILDV | MSTGSSSVGN | ATRIAHQLHQ | HRLKSLQPVV |
| 730 | 740 | 750 | 760 | 770 | 780 |
| SCGWLPPPAN | GHKEGLRYLE | GSVVRFSCNN | GYSLVGPESS | TCQADGKWSM | PTPECQPGRS |
| 790 | 800 | 810 | |||
| YTVLLSIIFG | GLAIVALISI | IYMMLHRRRK | SNMTMWSSQP |