Q9DB34
Gene name |
Chmp2a |
Protein name |
Charged multivesicular body protein 2a |
Names |
Chromatin-modifying protein 2a, CHMP2a, Vacuolar protein sorting-associated protein 2, mVps2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:68953 |
EC number |
|
Protein Class |
|
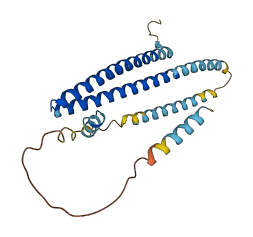
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1-144 (N-terminal α1-α4 domains) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9DB34
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9DB34-F1 | Predicted | AlphaFoldDB |
No variants for Q9DB34
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q9DB34 | |||||
No associated diseases with Q9DB34
2 regional properties for Q9DB34
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | High mobility group box domain | 49 - 119 | IPR009071-1 |
| domain | High mobility group box domain | 154 - 220 | IPR009071-2 |
Functions
14 GO annotations of cellular component
| Name | Definition |
|---|---|
| amphisome membrane | Any membrane that is part of an amphisome. |
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| ESCRT III complex | A complex with membrane scission activity that plays a major role in many processes where membranes are remodelled - including endosomal transport (vesicle budding), nuclear envelope organisation (membrane closure, mitotic bridge cleavage), and cytokinesis (abscission). |
| kinetochore | A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules. |
| kinetochore microtubule | Any of the spindle microtubules that attach to the kinetochores of chromosomes by their plus ends, and maneuver the chromosomes during mitotic or meiotic chromosome segregation. |
| lysosomal membrane | The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm. |
| membrane coat | Any of several different proteinaceous coats that can associate with membranes. Membrane coats include those formed by clathrin plus an adaptor complex, the COPI and COPII complexes, and possibly others. They are found associated with membranes on many vesicles as well as other membrane features such as pits and perhaps tubules. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| multivesicular body | A type of endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body membrane | The lipid bilayer surrounding a multivesicular body. |
| nuclear envelope | The double lipid bilayer enclosing the nucleus and separating its contents from the rest of the cytoplasm; includes the intermembrane space, a gap of width 20-40 nm (also called the perinuclear space). |
| nuclear pore | A protein complex providing a discrete opening in the nuclear envelope of a eukaryotic cell, where the inner and outer nuclear membranes are joined. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| phosphatidylcholine binding | Binding to a phosphatidylcholine, a glycophospholipid in which a phosphatidyl group is esterified to the hydroxyl group of choline. |
| protein domain specific binding | Binding to a specific domain of a protein. |
23 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome maturation | Removal of PI3P and Atg8/LC3 after the closure of the phagophore and before the fusion with the endosome/lysosome (e.g. mammals and insects) or vacuole (yeast), and that very likely destabilizes other Atg proteins and thus enables their efficient dissociation and recycling. |
| endosome transport via multivesicular body sorting pathway | The directed movement of substances from endosomes to lysosomes or vacuoles by a pathway in which molecules are sorted into multivesicular bodies, which then fuse with the target compartment. |
| establishment of protein localization | The directed movement of a protein to a specific location. |
| exit from mitosis | The cell cycle transition where a cell leaves M phase and enters a new G1 phase. M phase is the part of the mitotic cell cycle during which mitosis and cytokinesis take place. |
| late endosome to lysosome transport | The directed movement of substances from late endosome to lysosome. |
| late endosome to vacuole transport | The directed movement of substances from late endosomes to the vacuole. In yeast, after transport to the prevacuolar compartment, endocytic content is delivered to the late endosome and on to the vacuole. This pathway is analogous to endosome to lysosome transport. |
| membrane invagination | The infolding of a membrane. |
| midbody abscission | The process by which the midbody, the cytoplasmic bridge that connects the two prospective daughter cells, is severed at the end of mitotic cytokinesis, resulting in two separate daughter cells. |
| mitotic metaphase plate congression | The cell cycle process in which chromosomes are aligned at the metaphase plate, a plane halfway between the poles of the mitotic spindle, during mitosis. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| negative regulation of centriole elongation | Any process that stops, prevents or reduces the frequency, rate or extent of centriole elongation. |
| nuclear membrane reassembly | The reformation of the nuclear membranes following their breakdown in the context of a normal process. |
| nucleus organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the nucleus. |
| plasma membrane repair | The resealing of a cell plasma membrane after cellular wounding due to, for instance, mechanical stress. |
| positive regulation of exosomal secretion | Any process that activates or increases the frequency, rate or extent of exosomal secretion. |
| protein homooligomerization | The process of creating protein oligomers, compounds composed of a small number, usually between three and ten, of identical component monomers. Oligomers may be formed by the polymerization of a number of monomers or the depolymerization of a large protein polymer. |
| protein polymerization | The process of creating protein polymers, compounds composed of a large number of component monomers; polymeric proteins may be made up of different or identical monomers. Polymerization occurs by the addition of extra monomers to an existing poly- or oligomeric protein. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| regulation of centrosome duplication | Any process that modulates the frequency, rate or extent of centrosome duplication. Centrosome duplication is the replication of a centrosome, a structure comprised of a pair of centrioles and peri-centriolar material from which a microtubule spindle apparatus is organized. |
| regulation of mitotic spindle assembly | Any process that modulates the frequency, rate or extent of mitotic spindle assembly. |
| ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | The chemical reactions and pathways resulting in the breakdown of a protein or peptide covalently tagged with ubiquitin, via the multivesicular body (MVB) sorting pathway; ubiquitin-tagged proteins are sorted into MVBs, and delivered to a lysosome/vacuole for degradation. |
| viral budding from plasma membrane | A viral budding that starts with formation of a membrane curvature in the host plasma membrane. |
| viral budding via host ESCRT complex | Viral budding which uses a host ESCRT protein complex, or complexes, to mediate the budding process. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5ZHN1 | CHMP2A | Charged multivesicular body protein 2a | Gallus gallus (Chicken) | SS |
| O43633 | CHMP2A | Charged multivesicular body protein 2a | Homo sapiens (Human) | EV |
| Q9CQ10 | Chmp3 | Charged multivesicular body protein 3 | Mus musculus (Mouse) | SS |
| Q9CQD4 | Chmp1b2 | Charged multivesicular body protein 1b-2 | Mus musculus (Mouse) | PR |
| Q99LU0 | Chmp1b1 | Charged multivesicular body protein 1b-1 | Mus musculus (Mouse) | PR |
| Q9SKI2 | VPS2.1 | Vacuolar protein sorting-associated protein 2 homolog 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q6DFS6 | chmp2a | Charged multivesicular body protein 2a | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q7ZW25 | chmp2a | Charged multivesicular body protein 2a | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDLLFGRRKT | PEELLRQNQR | ALNRAMRELD | RERQKLETQE | KKIIADIKKM | AKQGQMDAVR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IMAKDLVRTR | RYVRKFVLMR | ANIQAVSLKI | QTLKSNNSMA | QAMKGVTKAM | GTMNRQLKLP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QIQKIMMEFE | RQAEIMDMKE | EMMNDAIDDA | MGDEEDEEES | DAVVSQVLDE | LGLSLTDELS |
| 190 | 200 | 210 | 220 | ||
| NLPSTGGSLS | VAAGGKKAEA | TASALADADA | DLEERLKNLR | RD |