Q9DAK3
Gene name |
Rhobtb1 |
Protein name |
Rho-related BTB domain-containing protein 1 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:69288 |
EC number |
|
Protein Class |
|
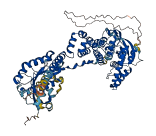
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9DAK3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9DAK3-F1 | Predicted | AlphaFoldDB |
30 variants for Q9DAK3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389097476 | 9 | R>Q | No | EVA | |
| rs3389071339 | 32 | C>Y | No | EVA | |
| rs3389071334 | 47 | A>T | No | EVA | |
| rs3389105920 | 67 | E>D | No | EVA | |
| rs3389048183 | 97 | Y>* | No | EVA | |
| rs3400909104 | 107 | F>L | No | EVA | |
| rs3389101810 | 113 | N>K | No | EVA | |
| rs3389104639 | 132 | R>Q | No | EVA | |
| rs3389092551 | 152 | V>I | No | EVA | |
| rs3389104602 | 171 | P>L | No | EVA | |
| rs3389105976 | 184 | P>Q | No | EVA | |
| rs3389078272 | 215 | Q>H | No | EVA | |
| rs29801653 | 275 | E>Q | No | EVA | |
| rs3389092585 | 306 | W>* | No | EVA | |
| rs235332736 | 319 | F>C | No | EVA | |
| rs3389109747 | 352 | L>Q | No | EVA | |
| rs237965775 | 385 | R>W | No | EVA | |
| rs1132202381 | 409 | P>R | No | EVA | |
| rs3389097468 | 457 | F>Y | No | EVA | |
| rs3389104746 | 491 | D>G | No | EVA | |
| rs3389048156 | 511 | A>G | No | EVA | |
| rs3389097442 | 568 | T>I | No | EVA | |
| rs3389071311 | 581 | E>K | No | EVA | |
| rs3389101775 | 599 | Y>* | No | EVA | |
| rs3400140264 | 600 | L>F | No | EVA | |
| rs3389048215 | 621 | T>I | No | EVA | |
| rs3401283576 | 637 | K>* | No | EVA | |
| rs3389104588 | 650 | W>R | No | EVA | |
| rs3389105917 | 652 | P>R | No | EVA | |
| rs3389071380 | 654 | W>* | No | EVA |
No associated diseases with Q9DAK3
3 regional properties for Q9DAK3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | BTB/POZ domain | 266 - 456 | IPR000210-1 |
| domain | BTB/POZ domain | 475 - 582 | IPR000210-2 |
| domain | Small GTP-binding protein domain | 14 - 150 | IPR005225 |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| cortical cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures in the cell cortex, i.e. just beneath the plasma membrane. |
| engulfment of apoptotic cell | The removal of the apoptotic cell by phagocytosis, by a neighboring cell or by a phagocyte. |
| establishment or maintenance of cell polarity | Any cellular process that results in the specification, formation or maintenance of anisotropic intracellular organization or cell growth patterns. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| small GTPase mediated signal transduction | The series of molecular signals in which a small monomeric GTPase relays a signal. |
36 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P61585 | RHOA | Transforming protein RhoA | Bos taurus (Bovine) | PR |
| Q1RMJ6 | RHOC | Rho-related GTP-binding protein RhoC | Bos taurus (Bovine) | PR |
| Q17QI8 | Rhov | Rho-related GTP-binding protein RhoV | Bos taurus (Bovine) | PR |
| Q2HJ68 | RND1 | Rho-related GTP-binding protein Rho6 | Bos taurus (Bovine) | PR |
| Q3ZBW5 | RHOB | Rho-related GTP-binding protein RhoB | Bos taurus (Bovine) | PR |
| Q9PSX7 | RHOC | Rho-related GTP-binding protein RhoC | Gallus gallus (Chicken) | PR |
| P48148 | Rho1 | Ras-like GTP-binding protein Rho1 | Drosophila melanogaster (Fruit fly) | PR |
| Q96L33 | RHOV | Rho-related GTP-binding protein RhoV | Homo sapiens (Human) | PR |
| P62745 | RHOB | Rho-related GTP-binding protein RhoB | Homo sapiens (Human) | PR |
| P08134 | RHOC | Rho-related GTP-binding protein RhoC | Homo sapiens (Human) | PR |
| Q92730 | RND1 | Rho-related GTP-binding protein Rho6 | Homo sapiens (Human) | PR |
| P61587 | RND3 | Rho-related GTP-binding protein RhoE | Homo sapiens (Human) | PR |
| P61586 | RHOA | Transforming protein RhoA | Homo sapiens (Human) | PR |
| O94955 | RHOBTB3 | Rho-related BTB domain-containing protein 3 | Homo sapiens (Human) | EV |
| O94844 | RHOBTB1 | Rho-related BTB domain-containing protein 1 | Homo sapiens (Human) | PR |
| Q9CTN4 | Rhobtb3 | Rho-related BTB domain-containing protein 3 | Mus musculus (Mouse) | SS |
| P61588 | Rnd3 | Rho-related GTP-binding protein RhoE | Mus musculus (Mouse) | PR |
| P62746 | Rhob | Rho-related GTP-binding protein RhoB | Mus musculus (Mouse) | PR |
| Q62159 | Rhoc | Rho-related GTP-binding protein RhoC | Mus musculus (Mouse) | PR |
| Q8VDU1 | Rhov | Rho-related GTP-binding protein RhoV | Mus musculus (Mouse) | PR |
| Q9QUI0 | Rhoa | Transforming protein RhoA | Mus musculus (Mouse) | PR |
| Q8BLR7 | Rnd1 | Rho-related GTP-binding protein Rho6 | Mus musculus (Mouse) | PR |
| O77683 | RND3 | Rho-related GTP-binding protein RhoE | Sus scrofa (Pig) | PR |
| Q6SA80 | Rnd3 | Rho-related GTP-binding protein RhoE | Rattus norvegicus (Rat) | PR |
| P62747 | Rhob | Rho-related GTP-binding protein RhoB | Rattus norvegicus (Rat) | PR |
| Q9Z1Y0 | Rhov | Rho-related GTP-binding protein RhoV | Rattus norvegicus (Rat) | PR |
| P61589 | Rhoa | Transforming protein RhoA | Rattus norvegicus (Rat) | PR |
| Q9SSX0 | RAC1 | Rac-like GTP-binding protein 1 | Oryza sativa subsp japonica (Rice) | PR |
| Q6Z808 | RAC3 | Rac-like GTP-binding protein 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q68Y52 | RAC2 | Rac-like GTP-binding protein 2 | Oryza sativa subsp japonica (Rice) | PR |
| Q67VP4 | RAC4 | Rac-like GTP-binding protein 4 | Oryza sativa subsp japonica (Rice) | PR |
| Q22038 | rho-1 | Ras-like GTP-binding protein rhoA | Caenorhabditis elegans | PR |
| Q9SU67 | ARAC8 | Rac-like GTP-binding protein ARAC8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82480 | ARAC7 | Rac-like GTP-binding protein ARAC7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38903 | ARAC2 | Rac-like GTP-binding protein ARAC2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82481 | ARAC10 | Rac-like GTP-binding protein ARAC10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDSDMDYERP | NVETIKCVVV | GDNAVGKTRL | ICARACNTTL | TQYQLLATHV | PTVWAIDQYR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VCQEVLERSR | DVVDEVSISL | RLWDTFGDHH | KDRRFAYGRS | DVVVLCFSIA | NPNSLNHVKT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MWYQEIKHFC | PRTPVVLVGC | QLDLRYADLE | AVNRARRPLA | RPIKRGDILP | PEKGREVAKE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LGIPYYETSV | FDQFGIKDVF | DNAIRAALIS | RRHLQFWKSH | LKKVQKPLLQ | APFLPPKAPP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PVIKVPECPS | AGTSDAACLL | DNPLCADVLF | VLHDEEHIFA | HRIYLATSSS | KFYDLFLMEC |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EESPCWGGGA | GEEVPCRDFQ | GRTQSLGSAE | EGKEGPQRTP | QADPGASSGQ | DLPESLALQM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EASGSEGHAL | SGWSKGFVSM | HREVRVNPIS | KRVGPVTVVR | LDPSMQSGPF | RTLLRFLYSG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| QLDEKEKDLL | GLAQMAEVLE | MFDLRMMVEN | IMNKEAFMNQ | EITKAFHVRK | ANRIKECLSK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GTFSDVTFTL | DDGAISAHKP | LLICSCEWMA | AMFGGSFVES | ANREVHLPNI | NKMSMQAVLE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| YLYTKQLSPN | LDLDPLELIA | LANRFCLTHL | VALVEQHAVQ | ELTKAAVSGV | SIDGEVLSYL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| ELAQFHNANQ | LAAWCLHHIC | TNYNSVCSKF | RKEIKSKSAD | NQEYFERHRW | PPVWYLKEED |
| 670 | 680 | 690 | |||
| HYQRVKRERE | KEDLALNKHH | SRRKWCFWHS | SPAVA |