Q9D5D8
Gene name |
Cdyl2 |
Protein name |
Chromodomain Y-like protein 2 |
Names |
CDY-like 2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:75796 |
EC number |
|
Protein Class |
|
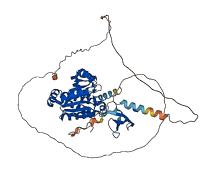
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9D5D8
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9D5D8-F1 | Predicted | AlphaFoldDB |
27 variants for Q9D5D8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389012029 | 16 | R>S | No | EVA | |
| rs3389015178 | 60 | L>* | No | EVA | |
| rs3388964324 | 82 | R>C | No | EVA | |
| rs3389014574 | 92 | R>W | No | EVA | |
| rs3389015244 | 96 | P>A | No | EVA | |
| rs3389007855 | 100 | K>I | No | EVA | |
| rs3388984374 | 109 | V>I | No | EVA | |
| rs241012459 | 114 | S>P | No | EVA | |
| rs3389014320 | 118 | K>N | No | EVA | |
| rs3388995396 | 119 | G>R | No | EVA | |
| rs3389019825 | 120 | S>E | No | EVA | |
| rs251683313 | 163 | D>E | No | EVA | |
| rs3389001582 | 232 | R>SE* | No | EVA | |
| rs3389014360 | 234 | R>G | No | EVA | |
| rs3389019857 | 253 | K>N | No | EVA | |
| rs3399697624 | 256 | G>C | No | EVA | |
| rs3389001543 | 265 | Q>R | No | EVA | |
| rs3388995395 | 273 | T>I | No | EVA | |
| rs3389012195 | 287 | A>T | No | EVA | |
| rs3389019812 | 300 | V>M | No | EVA | |
| rs3389006651 | 302 | S>N | No | EVA | |
| rs3389019862 | 330 | E>D | No | EVA | |
| rs3388995313 | 338 | A>S | No | EVA | |
| rs3388984350 | 378 | T>I | No | EVA | |
| rs3389012227 | 442 | V>M | No | EVA | |
| rs3389015186 | 452 | V>* | No | EVA | |
| rs3389006608 | 454 | E>G | No | EVA |
No associated diseases with Q9D5D8
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| catalytic activity | Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic. |
| methylated histone binding | Binding to a histone in which a residue has been modified by methylation. |
| transcription corepressor activity | A transcription coregulator activity that represses or decreases the transcription of specific gene sets via binding to a DNA-bound DNA-binding transcription factor, either on its own or as part of a complex. Corepressors often act by altering chromatin structure and modifications. For example, one class of transcription corepressors modifies chromatin structure through covalent modification of histones. A second class remodels the conformation of chromatin in an ATP-dependent fashion. A third class modulates interactions of DNA-bound DNA-binding transcription factors with other transcription coregulators. |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q99549 | MPHOSPH8 | M-phase phosphoprotein 8 | Homo sapiens (Human) | PR |
| P83916 | CBX1 | Chromobox protein homolog 1 | Homo sapiens (Human) | PR |
| Q9Y232 | CDYL | Chromodomain Y-like protein | Homo sapiens (Human) | PR |
| Q8N8U2 | CDYL2 | Chromodomain Y-like protein 2 | Homo sapiens (Human) | PR |
| P83917 | Cbx1 | Chromobox protein homolog 1 | Mus musculus (Mouse) | PR |
| Q9WTK2 | Cdyl | Chromodomain Y-like protein | Mus musculus (Mouse) | PR |
| Q6AYK9 | Cdyl | Chromodomain Y-like protein | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASGDLYEVE | RIVDKRKNKK | GKWEYLIRWK | GYGSTEDTWE | PEHHLLHCEE | FIDEFNGLHL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SKDKRVKSGK | QAGASKLLRD | ARGLPVERLS | HRPLEPGKSK | PSSHKRKRVN | SPLSRPKKGS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SGKAPDRATK | TVSYRTTPSG | LQIMPLKKAQ | NGLENGDAGS | EKDESHFGNG | SHQPDLELND |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QLGEQEASDC | DGTHSALVEN | GVGSALTNGG | LNLHSPVKRK | LETEKDYVFD | KRLRYSVRQN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ESNCRFRDIV | VRKEEGFTHI | LLSSQTSDNN | ALTPEIMKEV | RRALCNAATD | DSKLLLLSAV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GSVFCSGLDY | SYLIGRLSSD | RRKESTRIAE | AIRDFVKAFI | QFKKPIVVAI | NGPALGLGAS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ILPLCDIVWA | SEKAWFQTPY | ATIRLTPAGC | SSYTFPQILG | VALANEMLFC | GRKLTAQEAC |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SRGLVSQVFW | PTTFSQEVML | RVKEMASCSA | VVLEESKCLV | RSFLKSVLEE | VNEKECVMLK |
| 490 | 500 | ||||
| QLWSSSKGLD | SLFSYLQDKI | YEV |