Q9CXC9
Gene name |
Etv5 |
Protein name |
ETS translocation variant 5 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:104156 |
EC number |
|
Protein Class |
|
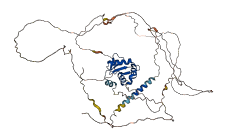
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
367-452 (ETS domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9CXC9
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9CXC9-F1 | Predicted | AlphaFoldDB |
31 variants for Q9CXC9
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389405689 | 6 | D>V | No | EVA | |
| rs3406822670 | 79 | V>E | No | EVA | |
| rs3407067851 | 80 | L>H | No | EVA | |
| rs3406663386 | 81 | H>D | No | EVA | |
| rs3389419335 | 81 | H>R | No | EVA | |
| rs3389405647 | 82 | A>T | No | EVA | |
| rs3406904734 | 84 | P>L | No | EVA | |
| rs3389419332 | 88 | I>V | No | EVA | |
| rs3389412193 | 103 | S>I | No | EVA | |
| rs3389324572 | 103 | S>R | No | EVA | |
| rs3389368962 | 112 | Y>* | No | EVA | |
| rs3389368927 | 114 | E>K | No | EVA | |
| rs3406800166 | 116 | C>G | No | EVA | |
| rs3389324517 | 182 | P>L | No | EVA | |
| rs3389414703 | 183 | E>G | No | EVA | |
| rs3389368901 | 221 | Q>R | No | EVA | |
| rs3389402451 | 240 | R>H | No | EVA | |
| rs3389405696 | 242 | S>I | No | EVA | |
| rs3405860482 | 266 | Y>S | No | EVA | |
| rs3389402751 | 273 | H>Y | No | EVA | |
| rs3389324573 | 301 | A>V | No | EVA | |
| rs3389377181 | 324 | G>* | No | EVA | |
| rs3389402458 | 345 | L>Q | No | EVA | |
| rs3389412187 | 353 | P>R | No | EVA | |
| rs3389388006 | 368 | L>M | No | EVA | |
| rs3389357949 | 381 | P>L | No | EVA | |
| rs3389402493 | 390 | T>S | No | EVA | |
| rs3389377094 | 435 | M>V | No | EVA | |
| rs3389368891 | 468 | K>T | No | EVA | |
| rs3389402721 | 483 | L>Q | No | EVA | |
| rs228076610 | 505 | T>A | No | EVA |
No associated diseases with Q9CXC9
6 regional properties for Q9CXC9
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | WW domain | 2389 - 2422 | IPR001202 |
| domain | SET domain | 1550 - 1673 | IPR001214 |
| domain | Post-SET domain | 1674 - 1690 | IPR003616 |
| domain | AWS domain | 1494 - 1549 | IPR006560 |
| domain | Set2 Rpb1 interacting domain | 2468 - 2556 | IPR013257 |
| domain | SETD2/Set2, SET domain | 1549 - 1690 | IPR044437 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
11 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to oxidative stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| locomotory behavior | The specific movement from place to place of an organism in response to external or internal stimuli. Locomotion of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions. |
| male germ-line stem cell asymmetric division | The self-renewing division of a germline stem cell in the male gonad, to produce a daughter stem cell and a daughter germ cell, which will divide to form the male gametes. |
| neuromuscular synaptic transmission | The process of synaptic transmission from a neuron to a muscle, across a synapse. |
| positive regulation of DNA-templated transcription | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| positive regulation of glial cell proliferation | Any process that activates or increases the rate or extent of glial cell proliferation. |
| positive regulation of neuron differentiation | Any process that activates or increases the frequency, rate or extent of neuron differentiation. |
| regulation of branching involved in mammary gland duct morphogenesis | Any process that modulates the rate, frequency, or extent of branching involved in mammary gland duct morphogenesis. |
| regulation of synapse organization | Any process that modulates the physical form of a synapse, the junction between a neuron and a target (neuron, muscle, or secretory cell). |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| skeletal muscle acetylcholine-gated channel clustering | The accumulation of acetylcholine-gated cation channels in a narrow, central region of muscle fibers, in apposition to nerve terminals. |
39 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A1A4L6 | ETS2 | Protein C-ets-2 | Bos taurus (Bovine) | SS |
| Q2KIC2 | ETV1 | ETS translocation variant 1 | Bos taurus (Bovine) | SS |
| P15062 | ETS1 | Transforming protein p68/c-ets-1 | Gallus gallus (Chicken) | SS |
| P10157 | ETS2 | Protein C-ets-2 | Gallus gallus (Chicken) | SS |
| Q90837 | ERG | Transcriptional regulator Erg | Gallus gallus (Chicken) | SS |
| A2T762 | ETV3 | ETS translocation variant 3 | Pan troglodytes (Chimpanzee) | PR |
| Q04688 | Ets97D | DNA-binding protein Ets97D | Drosophila melanogaster (Fruit fly) | PR |
| P50548 | ERF | ETS domain-containing transcription factor ERF | Homo sapiens (Human) | PR |
| P41970 | ELK3 | ETS domain-containing protein Elk-3 | Homo sapiens (Human) | SS |
| P43268 | ETV4 | ETS translocation variant 4 | Homo sapiens (Human) | EV |
| Q06546 | GABPA | GA-binding protein alpha chain | Homo sapiens (Human) | SS |
| P28324 | ELK4 | ETS domain-containing protein Elk-4 | Homo sapiens (Human) | EV |
| P15036 | ETS2 | Protein C-ets-2 | Homo sapiens (Human) | EV |
| P41162 | ETV3 | ETS translocation variant 3 | Homo sapiens (Human) | PR |
| P14921 | ETS1 | Protein C-ets-1 | Homo sapiens (Human) | EV |
| P50549 | ETV1 | ETS translocation variant 1 | Homo sapiens (Human) | EV |
| P11308 | ERG | Transcriptional regulator ERG | Homo sapiens (Human) | EV |
| P19419 | ELK1 | ETS domain-containing protein Elk-1 | Homo sapiens (Human) | EV |
| P41161 | ETV5 | ETS translocation variant 5 | Homo sapiens (Human) | SS |
| P15037 | Ets2 | Protein C-ets-2 | Mus musculus (Mouse) | SS |
| P41158 | Elk4 | ETS domain-containing protein Elk-4 | Mus musculus (Mouse) | PR |
| P41164 | Etv1 | ETS translocation variant 1 | Mus musculus (Mouse) | SS |
| P41969 | Elk1 | ETS domain-containing protein Elk-1 | Mus musculus (Mouse) | PR |
| P41971 | Elk3 | ETS domain-containing protein Elk-3 | Mus musculus (Mouse) | EV |
| Q00422 | Gabpa | GA-binding protein alpha chain | Mus musculus (Mouse) | EV |
| P97360 | Etv6 | Transcription factor ETV6 | Mus musculus (Mouse) | EV |
| Q8VDK3 | Elf5 | ETS-related transcription factor Elf-5 | Mus musculus (Mouse) | SS |
| Q60775 | Elf1 | ETS-related transcription factor Elf-1 | Mus musculus (Mouse) | PR |
| Q9Z2U4 | Elf4 | ETS-related transcription factor Elf-4 | Mus musculus (Mouse) | PR |
| Q9JHC9 | Elf2 | ETS-related transcription factor Elf-2 | Mus musculus (Mouse) | PR |
| Q9WTP3 | Spdef | SAM pointed domain-containing Ets transcription factor | Mus musculus (Mouse) | PR |
| P70459 | Erf | ETS domain-containing transcription factor ERF | Mus musculus (Mouse) | PR |
| P81270 | Erg | Transcriptional regulator ERG | Mus musculus (Mouse) | SS |
| P28322 | Etv4 | ETS translocation variant 4 | Mus musculus (Mouse) | SS |
| P27577 | Ets1 | Protein C-ets-1 | Mus musculus (Mouse) | EV |
| P41156 | Ets1 | Protein C-ets-1 | Rattus norvegicus (Rat) | SS |
| A4GTP4 | Elk1 | ETS domain-containing protein Elk-1 | Rattus norvegicus (Rat) | PR |
| Q9PUQ1 | etv4 | ETS translocation variant 4 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A3FEM2 | fev | Protein FEV | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDGFCDQQVP | FMVPGKSRSE | DCRGRPLIDR | KRKFVDTDLA | HDSEELFQDL | SQLQEAWLAE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AQVPDDEQFV | PDFQSDNLVL | HAPPPTKIKR | ELHSPSSELS | SCSHEQALGA | KYGEKCLYNY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| CAYDRKPPSG | FKPLTPPATP | LSPTHQNSLF | PPPQATLPTS | GLTPGAGPVQ | GVGPAPTPHS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LPEPGSQQQT | FAVPRPPHQP | LQMPKMMPES | QYPSEQRFQR | QLSEPSHPFP | PQSGVPGDSR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PSYHRQMSEP | IVPAAPPPLQ | GFKQEYHDPL | YEHGVPGMPG | PPAHGFQSPM | GIKQEPRDYC |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ADSEVPNCQS | SYMRGGYFSS | SHEGFPYEKD | PRLYFDDTCV | VPERLEGKVK | QEPTMYREGP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PYQRRGSLQL | WQFLVTLLDD | PANAHFIAWT | GRGMEFKLIE | PEEVARRWGI | QKNRPAMNYD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KLSRSLRYYY | EKGIMQKVAG | ERYVYKFVCD | PDALFSMAFP | DNQRPFLKAE | SECPLNEEDT |
| 490 | 500 | ||||
| LPLTHFEDNP | AYLLDMDRCS | SLPYTEGFAY |