Q9C646
Gene name |
RXW24L (At1g58410, F9K23.6, X7J.10) |
Protein name |
Probable disease resistance protein RXW24L |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G58410 |
EC number |
|
Protein Class |
|
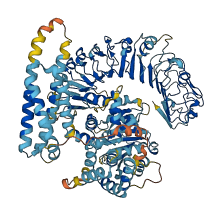
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q9C646
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9C646-F1 | Predicted | AlphaFoldDB |
223 variants for Q9C646
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_1_21704239_A_C | 6 | F>C | No | 1000Genomes | |
| ENSVATH01395095 | 6 | F>L | No | 1000Genomes | |
| ENSVATH04994732 | 13 | D>N | No | 1000Genomes | |
| ENSVATH13633292 | 19 | Y>H | No | 1000Genomes | |
| ENSVATH01395092 | 20 | D>E | No | 1000Genomes | |
| ENSVATH13633291 | 22 | F>S | No | 1000Genomes | |
| ENSVATH01395091 | 23 | K>N | No | 1000Genomes | |
| tmp_1_21704150_G_C | 36 | L>V | No | 1000Genomes | |
| tmp_1_21704124_T_G | 44 | K>N | No | 1000Genomes | |
| ENSVATH04994729 | 46 | A>T | No | 1000Genomes | |
| ENSVATH04994728 | 52 | I>V | No | 1000Genomes | |
| ENSVATH04994725 | 54 | E>K | No | 1000Genomes | |
| ENSVATH01395085 | 61 | E>V | No | 1000Genomes | |
| ENSVATH01395084 | 65 | D>E | No | 1000Genomes | |
| ENSVATH13633286 | 78 | I>L | No | 1000Genomes | |
| ENSVATH13633285 | 79 | L>H | No | 1000Genomes | |
| ENSVATH04994722 | 85 | M>I | No | 1000Genomes | |
| ENSVATH04994723 | 85 | M>K | No | 1000Genomes | |
| ENSVATH04994721 | 95 | R>* | No | 1000Genomes | |
| ENSVATH00104848 | 101 | M>V | No | 1000Genomes | |
| ENSVATH04994720 | 104 | R>M | No | 1000Genomes | |
| ENSVATH01395077 | 110 | I>T | No | 1000Genomes | |
| ENSVATH04994719 | 111 | G>E | No | 1000Genomes | |
| ENSVATH04994718 | 114 | S>R | No | 1000Genomes | |
| ENSVATH04994716 | 120 | V>G | No | 1000Genomes | |
| ENSVATH14350974 | 126 | S>I | No | 1000Genomes | |
| tmp_1_21703855_G_C | 134 | T>S | No | 1000Genomes | |
| ENSVATH04994714 | 158 | S>Y | No | 1000Genomes | |
| tmp_1_21703780_T_A | 159 | E>V | No | 1000Genomes | |
| ENSVATH04994712 | 165 | M>I | No | 1000Genomes | |
| ENSVATH01395074 | 165 | M>L | No | 1000Genomes | |
| tmp_1_21703762_A_G | 165 | M>T | No | 1000Genomes | |
| ENSVATH01395072 | 171 | K>I | No | 1000Genomes | |
| tmp_1_21703735_C_T | 174 | G>E | No | 1000Genomes | |
| ENSVATH01395070 | 177 | V>L | No | 1000Genomes | |
| ENSVATH01395069 | 178 | E>G | No | 1000Genomes | |
| ENSVATH13633257 | 185 | V>F | No | 1000Genomes | |
| tmp_1_21703691_C_G | 189 | G>R | No | 1000Genomes | |
| tmp_1_21703681_C_G | 192 | G>A | No | 1000Genomes | |
| ENSVATH01395062 | 198 | L>V | No | 1000Genomes | |
| tmp_1_21703640_C_T | 206 | D>N | No | 1000Genomes | |
| ENSVATH13633254 | 220 | V>I | No | 1000Genomes | |
| ENSVATH04994705 | 228 | S>L | No | 1000Genomes | |
| ENSVATH04994706 | 228 | S>P | No | 1000Genomes | |
| ENSVATH13633252 | 238 | T>I | No | 1000Genomes | |
| ENSVATH04994704 | 244 | D>E | No | 1000Genomes | |
| ENSVATH04994703 | 247 | Q>H | No | 1000Genomes | |
| ENSVATH00104843 | 251 | E>K | No | 1000Genomes | |
| ENSVATH04994702 | 252 | A>G | No | 1000Genomes | |
| ENSVATH01395057 | 252 | A>S | No | 1000Genomes | |
| ENSVATH04994701 | 267 | T>S | No | 1000Genomes | |
| ENSVATH13633249 | 274 | I>M | No | 1000Genomes | |
| ENSVATH04994698 | 276 | K>N | No | 1000Genomes | |
| ENSVATH01395054 | 277 | E>K | No | 1000Genomes | |
| ENSVATH01395053 | 282 | L>V | No | 1000Genomes | |
| ENSVATH13633248 | 289 | P>A | No | 1000Genomes | |
| ENSVATH01395052 | 290 | K>E | No | 1000Genomes | |
| ENSVATH04994697 | 291 | K>N | No | 1000Genomes | |
| ENSVATH04994695 | 305 | A>P | No | 1000Genomes | |
| tmp_1_21703233_C_T | 308 | G>E | No | 1000Genomes | |
| tmp_1_21703228_T_C | 310 | T>A | No | 1000Genomes | |
| ENSVATH13633234 | 317 | P>R | No | 1000Genomes | |
| tmp_1_21703203_T_C | 318 | K>R | No | 1000Genomes | |
| ENSVATH13633233 | 330 | Q>* | No | 1000Genomes | |
| ENSVATH13633232 | 330 | Q>R | No | 1000Genomes | |
| tmp_1_21703162_T_C | 332 | I>V | No | 1000Genomes | |
| tmp_1_21703159_C_T | 333 | A>T | No | 1000Genomes | |
| ENSVATH01395035 | 342 | F>Y | No | 1000Genomes | |
| ENSVATH13633214 | 346 | E>K | No | 1000Genomes | |
| ENSVATH01395034 | 350 | N>D | No | 1000Genomes | |
| ENSVATH09345361 | 350 | N>K | No | 1000Genomes | |
| ENSVATH01395033 | 351 | M>L | No | 1000Genomes | |
| tmp_1_21702929_T_G | 353 | K>Q | No | 1000Genomes | |
| ENSVATH13633213 | 354 | K>E | No | 1000Genomes | |
| ENSVATH13633211 | 355 | M>I | No | 1000Genomes | |
| ENSVATH01395029 | 363 | S>P | No | 1000Genomes | |
| ENSVATH13633209 | 378 | T>K | No | 1000Genomes | |
| ENSVATH04994687 | 378 | T>S | No | 1000Genomes | |
| ENSVATH01395022 | 389 | I>S | No | 1000Genomes | |
| ENSVATH04994683 | 390 | G>R | No | 1000Genomes | |
| ENSVATH01395021 | 395 | E>K | No | 1000Genomes | |
| ENSVATH00104842 | 396 | R>T | No | 1000Genomes | |
| tmp_1_21702786_G_T | 400 | N>K | No | 1000Genomes | |
| tmp_1_21702787_T_G | 400 | N>T | No | 1000Genomes | |
| tmp_1_21702780_A_C | 402 | S>R | No | 1000Genomes | |
| ENSVATH13633206 | 405 | D>G | No | 1000Genomes | |
| ENSVATH01395020 | 410 | V>M | No | 1000Genomes | |
| ENSVATH04994682 | 412 | F>V | No | 1000Genomes | |
| tmp_1_21702710_G_A | 426 | L>F | No | 1000Genomes | |
| tmp_1_21702695_C_T | 431 | E>K | No | 1000Genomes | |
| ENSVATH01395018 | 448 | G>E | No | 1000Genomes | |
| tmp_1_21702629_T_A | 453 | R>W | No | 1000Genomes | |
| ENSVATH00104839 | 454 | R>H | No | 1000Genomes | |
| ENSVATH04994678 | 455 | Y>* | No | 1000Genomes | |
| ENSVATH04994679 | 455 | Y>N | No | 1000Genomes | |
| ENSVATH13633183 | 456 | D>N | No | 1000Genomes | |
| tmp_1_21702617_C_T | 457 | G>R | No | 1000Genomes | |
| ENSVATH14350969 | 460 | I>S | No | 1000Genomes | |
| ENSVATH00104838 | 463 | T>I | No | 1000Genomes | |
| ENSVATH00104837 | 468 | I>L | No | 1000Genomes | |
| tmp_1_21702581_C_T | 469 | E>K | No | 1000Genomes | |
| ENSVATH00104836 | 470 | E>D | No | 1000Genomes | |
| ENSVATH04994674 | 481 | R>W | No | 1000Genomes | |
| tmp_1_21702534_C_A | 484 | M>I | No | 1000Genomes | |
| ENSVATH00104835 | 484 | M>L | No | 1000Genomes | |
| ENSVATH00104834 | 489 | E>K | No | 1000Genomes | |
| ENSVATH01395015 | 492 | R>C | No | 1000Genomes | |
| ENSVATH04994673 | 494 | H>L | No | 1000Genomes | |
| tmp_1_21702506_G_A | 494 | H>Y | No | 1000Genomes | |
| tmp_1_21702498_C_T | 496 | M>I | No | 1000Genomes | |
| ENSVATH04994672 | 503 | F>L | No | 1000Genomes | |
| tmp_1_21702478_A_G | 503 | F>S | No | 1000Genomes | |
| ENSVATH01395014 | 504 | K>I | No | 1000Genomes | |
| tmp_1_21702476_T_G | 504 | K>Q | No | 1000Genomes | |
| ENSVATH01395014 | 504 | K>R | No | 1000Genomes | |
| ENSVATH04994671 | 508 | E>D | No | 1000Genomes | |
| ENSVATH01395013 | 512 | Q>H | No | 1000Genomes | |
| ENSVATH04994670 | 513 | I>T | No | 1000Genomes | |
| ENSVATH13633182 | 517 | H>D | No | 1000Genomes | |
| ENSVATH01395012 | 519 | P>T | No | 1000Genomes | |
| ENSVATH00104832 | 523 | P>A | No | 1000Genomes | |
| ENSVATH13633181 | 531 | R>* | No | 1000Genomes | |
| tmp_1_21702363_A_T | 541 | H>Q | No | 1000Genomes | |
| ENSVATH01395010 | 543 | E>D | No | 1000Genomes | |
| ENSVATH01395011 | 543 | E>Q | No | 1000Genomes | |
| tmp_1_21702351_A_C | 545 | Y>* | No | 1000Genomes | |
| ENSVATH01395009 | 549 | P>L | No | 1000Genomes | |
| ENSVATH13633180 | 552 | R>* | No | 1000Genomes | |
| ENSVATH13633179 | 554 | L>V | No | 1000Genomes | |
| ENSVATH00104831 | 558 | Y>S | No | 1000Genomes | |
| tmp_1_21702299_T_C | 563 | N>D | No | 1000Genomes | |
| tmp_1_21702297_A_T | 563 | N>K | No | 1000Genomes | |
| ENSVATH01395008 | 576 | V>L | No | 1000Genomes | |
| ENSVATH01395003 | 585 | V>I | No | 1000Genomes | |
| ENSVATH14350966 | 587 | A>G | No | 1000Genomes | |
| tmp_1_21702222_C_A,G | 588 | K>N | No | 1000Genomes | |
| ENSVATH01395002 | 589 | F>L | No | 1000Genomes | |
| ENSVATH01395001 | 591 | G>E | No | 1000Genomes | |
| ENSVATH13633176 | 597 | D>Y | No | 1000Genomes | |
| tmp_1_21702164_T_G | 608 | S>R | No | 1000Genomes | |
| ENSVATH04994665 | 612 | A>D | No | 1000Genomes | |
| ENSVATH01394994 | 619 | S>Y | No | 1000Genomes | |
| ENSVATH04994663 | 624 | L>I | No | 1000Genomes | |
| tmp_1_21702097_A_T | 630 | L>* | No | 1000Genomes | |
| ENSVATH14350965 | 631 | D>Y | No | 1000Genomes | |
| tmp_1_21702092_T_A | 632 | I>L | No | 1000Genomes | |
| ENSVATH13633163 | 633 | R>H | No | 1000Genomes | |
| ENSVATH04994661 | 635 | D>N | No | 1000Genomes | |
| ENSVATH13633162 | 636 | F>C | No | 1000Genomes | |
| ENSVATH13633160 | 639 | I>R | No | 1000Genomes | |
| ENSVATH04994659 | 642 | P>L | No | 1000Genomes | |
| ENSVATH13633157 | 647 | G>R | No | 1000Genomes | |
| ENSVATH14350944 | 649 | R>K | No | 1000Genomes | |
| ENSVATH04994657 | 660 | M>I | No | 1000Genomes | |
| ENSVATH04994656 | 661 | H>Y | No | 1000Genomes | |
| tmp_1_21701987_C_G | 667 | E>Q | No | 1000Genomes | |
| tmp_1_21701982_C_A | 668 | L>F | No | 1000Genomes | |
| tmp_1_21701972_C_G | 672 | E>Q | No | 1000Genomes | |
| tmp_1_21701941_G_T | 682 | T>K | No | 1000Genomes | |
| ENSVATH13633155 | 684 | S>N | No | 1000Genomes | |
| ENSVATH13633154 | 686 | S>I | No | 1000Genomes | |
| ENSVATH13633153 | 688 | E>D | No | 1000Genomes | |
| tmp_1_21701870_C_T | 706 | G>R | No | 1000Genomes | |
| ENSVATH13633147 | 708 | S>T | No | 1000Genomes | |
| ENSVATH00104827 | 712 | L>R | No | 1000Genomes | |
| ENSVATH14350942 | 713 | S>F | No | 1000Genomes | |
| tmp_1_21701849_A_T | 713 | S>T | No | 1000Genomes | |
| ENSVATH13633146 | 714 | A>T | No | 1000Genomes | |
| ENSVATH01394985 | 717 | C>R | No | 1000Genomes | |
| tmp_1_21701821_A_T | 722 | L>* | No | 1000Genomes | |
| ENSVATH01394984 | 723 | E>K | No | 1000Genomes | |
| ENSVATH01394983 | 724 | N>I | No | 1000Genomes | |
| ENSVATH01394982 | 725 | F>L | No | 1000Genomes | |
| ENSVATH04994653 | 727 | I>T | No | 1000Genomes | |
| tmp_1_21701802_C_T | 728 | M>I | No | 1000Genomes | |
| ENSVATH13633134 | 731 | A>T | No | 1000Genomes | |
| ENSVATH13633133 | 731 | A>V | No | 1000Genomes | |
| ENSVATH04994652 | 735 | R>K | No | 1000Genomes | |
| ENSVATH04994651 | 738 | E>K | No | 1000Genomes | |
| tmp_1_21701737_T_A | 750 | K>M | No | 1000Genomes | |
| tmp_1_21701731_G_C | 752 | T>S | No | 1000Genomes | |
| ENSVATH04994650 | 754 | S>R | No | 1000Genomes | |
| ENSVATH04994649 | 759 | R>M | No | 1000Genomes | |
| tmp_1_21701705_G_A | 761 | P>S | No | 1000Genomes | |
| tmp_1_21701679_G_T | 769 | H>Q | No | 1000Genomes | |
| ENSVATH04994646 | 794 | L>S | No | 1000Genomes | |
| tmp_1_21701602_T_G | 795 | K>T | No | 1000Genomes | |
| ENSVATH04994645 | 801 | Y>D | No | 1000Genomes | |
| tmp_1_21701578_G_A | 803 | S>F | No | 1000Genomes | |
| tmp_1_21701569_C_G | 806 | G>A | No | 1000Genomes | |
| ENSVATH13633127 | 806 | G>R | No | 1000Genomes | |
| ENSVATH13633126 | 812 | S>T | No | 1000Genomes | |
| ENSVATH13633125 | 813 | A>P | No | 1000Genomes | |
| ENSVATH13633114 | 813 | A>V | No | 1000Genomes | |
| ENSVATH04994644 | 817 | P>H | No | 1000Genomes | |
| tmp_1_21701537_G_A | 817 | P>S | No | 1000Genomes | |
| ENSVATH04994643 | 820 | R>L | No | 1000Genomes | |
| ENSVATH04994643 | 820 | R>P | No | 1000Genomes | |
| ENSVATH04994642 | 823 | A>D | No | 1000Genomes | |
| ENSVATH04994642 | 823 | A>V | No | 1000Genomes | |
| ENSVATH14350941 | 826 | E>K | No | 1000Genomes | |
| ENSVATH01394976 | 829 | E>V | No | 1000Genomes | |
| tmp_1_21701488_C_A | 833 | W>L | No | 1000Genomes | |
| ENSVATH01394975 | 837 | E>K | No | 1000Genomes | |
| ENSVATH13633105 | 838 | G>C | No | 1000Genomes | |
| ENSVATH01394974 | 841 | S>P | No | 1000Genomes | |
| tmp_1_21701461_C_A | 842 | R>L | No | 1000Genomes | |
| tmp_1_21701440_C_T | 849 | W>* | No | 1000Genomes | |
| ENSVATH01394972 | 857 | P>H | No | 1000Genomes | |
| ENSVATH04994640 | 861 | R>Q | No | 1000Genomes | |
| tmp_1_21701396_A_T | 864 | Y>N | No | 1000Genomes | |
| tmp_1_21701395_T_G | 864 | Y>S | No | 1000Genomes | |
| ENSVATH04994639 | 866 | L>I | No | 1000Genomes | |
| ENSVATH13633101 | 877 | E>D | No | 1000Genomes | |
| ENSVATH13633102 | 877 | E>K | No | 1000Genomes | |
| ENSVATH00104826 | 881 | E>K | No | 1000Genomes | |
| tmp_1_21701333_C_T | 885 | E>K | No | 1000Genomes | |
| ENSVATH04994637 | 887 | Y>D | No | 1000Genomes | |
| ENSVATH01394969 | 890 | Q>K | No | 1000Genomes | |
| tmp_1_21701314_T_C | 891 | N>S | No | 1000Genomes | |
| ENSVATH01394967 | 894 | F>S | No | 1000Genomes | |
| ENSVATH01394966 | 899 | S>F | No | 1000Genomes | |
| tmp_1_21701287_T_G | 900 | S>S | No | 1000Genomes |
No associated diseases with Q9C646
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| ADP binding | Binding to ADP, adenosine 5'-diphosphate. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| defense response | Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack. |
| response to other organism | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism. |
20 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| F1MHT9 | NLRC4 | NLR family CARD domain-containing protein 4 | Bos taurus (Bovine) | SS |
| O14727 | APAF1 | Apoptotic protease-activating factor 1 | Homo sapiens (Human) | EV |
| Q9NPP4 | NLRC4 | NLR family CARD domain-containing protein 4 | Homo sapiens (Human) | SS |
| Q3UP24 | Nlrc4 | NLR family CARD domain-containing protein 4 | Mus musculus (Mouse) | EV |
| F1M649 | Nlrc4 | NLR family CARD domain-containing protein 4 | Rattus norvegicus (Rat) | SS |
| O22727 | At1g61190 | Probable disease resistance protein At1g61190 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64790 | At1g61300 | Probable disease resistance protein At1g61300 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64973 | RPS5 | Disease resistance protein RPS5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P0C8S1 | RPP8L2 | Probable disease resistance RPP8-like protein 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P60838 | SUMM2 | Disease resistance protein SUMM2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38834 | RPP13L4 | Disease resistance RPP13-like protein 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39214 | RPM1 | Disease resistance protein RPM1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8W3K3 | At1g58400 | Putative disease resistance protein At1g58400 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FJB5 | RPP8L3 | Disease resistance RPP8-like protein 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRR4 | RPPL1 | Putative disease resistance RPP13-like protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9T048 | At4g27190 | Disease resistance protein At4g27190 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9XIF0 | At1g59780 | Putative disease resistance protein At1g59780 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64789 | At1g61310 | Probable disease resistance protein At1g61310 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q940K0 | UNI | Disease resistance protein UNI | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9I9H8 | apaf1 | Apoptotic protease-activating factor 1 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MELVSFGVEK | LWDRLSQEYD | QFKGVEDQVT | ELKSNLNLLK | SFLKDADAKK | HISEMVRHCV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EEIKDIVYDT | EDIIETFILK | EKVEMKRGIM | KRIKRFASTI | MDRRELASDI | GGISKRISKV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IQDMQSFGVQ | QIITDGSRSS | HPLQERQREM | RHTFSRDSEN | DFVGMEANVK | KLVGYLVEKD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DYQIVSLTGM | GGLGKTTLAR | QVFNHDVVKD | RFDGFAWVSV | SQEFTRISVW | QTILQNLTSK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ERKDEIQNMK | EADLHDDLFR | LLESSKTLIV | LDDIWKEEDW | DLIKPIFPPK | KGWKVLLTSR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TESIAMRGDT | TYISFKPKCL | SIPDSWTLFQ | SIAMPRKDTS | EFKVDEEMEN | MGKKMIKHCG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GLSLAVKVLG | GLLAAKYTLH | DWKRLSENIG | SHIVERTSGN | NSSIDHVLSV | SFEELPNYLK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| HCFLYLAHFP | EDHEIDVEKL | HYYWAAEGIS | ERRRYDGETI | RDTGDSYIEE | LVRRNMVISE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RDVMTSRFET | CRLHDMMREI | CLFKAKEENF | LQIVSNHSPT | SNPQTLGASR | RFVLHNPTTL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| HVERYKNNPK | LRSLVVVYDD | IGNRRWMLSG | SIFTRVKLLR | VLDLVQAKFK | GGKLPSDIGK |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LIHLRYLSLK | DAKVSHLPSS | LRNLVLLIYL | DIRTDFTDIF | VPNVFMGMRE | LRYLELPRFM |
| 670 | 680 | 690 | 700 | 710 | 720 |
| HEKTKLELSN | LEKLEALENF | STKSSSLEDL | RGMVRLRTLV | IILSEGTSLQ | TLSASVCGLR |
| 730 | 740 | 750 | 760 | 770 | 780 |
| HLENFKIMEN | AGVNRMGEER | MVLDFTYLKK | LTLSIEMPRL | PKIQHLPSHL | TVLDLSYCCL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| EEDPMPILEK | LLELKDLSLD | YLSFSGRKMV | CSAGGFPQLR | KLALDEQEEW | EEWIVEEGSM |
| 850 | 860 | 870 | 880 | 890 | |
| SRLHTLSIWS | STLKELPDGL | RFIYSLKNLI | MGKSWMERLS | ERGEEFYKVQ | NIPFIKFSS |