Q9BY43
Gene name |
CHMP4A |
Protein name |
Charged multivesicular body protein 4a |
Names |
Chromatin-modifying protein 4a, CHMP4a, SNF7 homolog associated with Alix-2, SNF7-1, hSnf-1, Vacuolar protein sorting-associated protein 32-1, Vps32-1, hVps32-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:29082 |
EC number |
|
Protein Class |
|
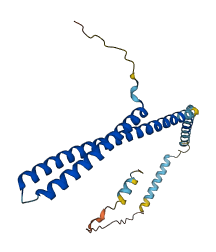
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1-144 (N-terminal α1-α4 domains) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

3 structures for Q9BY43
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3C3O | X-ray | 215 A | B | 210-222 | PDB |
| 5MK1 | X-ray | 250 A | E/F/H/K | 205-222 | PDB |
| AF-Q9BY43-F1 | Predicted | AlphaFoldDB |
334 variants for Q9BY43
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1355912999 CA389328571 |
3 | G>S | No |
ClinGen TOPMed gnomAD |
|
| rs1355912999 | 3 | G>S | No |
TOPMed gnomAD |
|
| rs1167943844 | 5 | G>C | No | TOPMed | |
|
CA389328557 rs1167943844 |
5 | G>C | No |
ClinGen TOPMed |
|
| rs144411001 | 6 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7129619 rs144411001 |
6 | R>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs779952199 | 7 | L>F | No |
ExAC TOPMed gnomAD |
|
|
rs779952199 CA7129617 |
7 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA389328542 rs1457849995 |
8 | F>L | No |
ClinGen gnomAD |
|
| rs1457849995 | 8 | F>L | No | gnomAD | |
| rs1404788543 | 9 | G>V | No | TOPMed | |
|
rs1404788543 CA389328530 |
9 | G>V | No |
ClinGen TOPMed |
|
|
CA7129616 rs533438540 |
10 | K>* | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| rs533438540 | 10 | K>* | No |
1000Genomes ExAC gnomAD |
|
|
rs765685878 CA7129581 |
11 | G>E | No |
ClinGen ExAC gnomAD |
|
| rs765685878 | 11 | G>E | No |
ExAC gnomAD |
|
| rs149642194 | 11 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
|
rs149642194 CA389328521 |
11 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs149642194 CA7129615 |
11 | G>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs149642194 | 11 | G>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs757811080 | 12 | K>Q | No |
ExAC TOPMed gnomAD |
|
|
CA7129580 rs757811080 |
12 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7129579 rs147406718 |
14 | E>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147406718 | 14 | E>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1594470213 | 17 | P>L | No | Ensembl | |
|
rs1594470213 CA389328320 |
17 | P>L | No |
ClinGen Ensembl |
|
| rs760234379 | 17 | P>S | No | ExAC | |
|
CA7129577 rs760234379 |
17 | P>S | No |
ClinGen ExAC |
|
| rs1375826151 | 18 | T>I | No | gnomAD | |
|
rs1375826151 CA389328312 |
18 | T>I | No |
ClinGen gnomAD |
|
| rs1308822959 | 19 | P>L | No | gnomAD | |
|
rs1308822959 CA389328306 |
19 | P>L | No |
ClinGen gnomAD |
|
| rs375389578 | 24 | Q>* | No |
1000Genomes ESP ExAC gnomAD |
|
|
CA7129574 rs375389578 |
24 | Q>* | No |
ClinGen 1000Genomes ESP ExAC gnomAD |
|
| rs375389578 | 24 | Q>E | No |
1000Genomes ESP ExAC gnomAD |
|
|
CA7129575 rs375389578 |
24 | Q>E | No |
ClinGen 1000Genomes ESP ExAC gnomAD |
|
| rs1226800374 | 28 | E>D | No | gnomAD | |
|
CA389328243 rs1226800374 |
28 | E>D | No |
ClinGen gnomAD |
|
| rs774307454 | 29 | T>I | No |
ExAC TOPMed gnomAD |
|
|
CA389328237 rs774307454 |
29 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774307454 CA7129573 |
29 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs774307454 | 29 | T>R | No |
ExAC TOPMed gnomAD |
|
|
rs146309316 CA7129572 |
30 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs146309316 | 30 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1011892227 | 33 | L>R | No | gnomAD | |
|
rs1011892227 CA257896935 |
33 | L>R | No |
ClinGen gnomAD |
|
| rs1387575321 | 33 | L>V | No | TOPMed | |
|
rs1387575321 CA389328213 |
33 | L>V | No |
ClinGen TOPMed |
|
|
CA389328208 rs1264841190 |
34 | I>F | No |
ClinGen TOPMed |
|
| rs1264841190 | 34 | I>F | No | TOPMed | |
| rs776846184 | 38 | E>K | No | ExAC | |
|
CA7129569 rs776846184 |
38 | E>K | No |
ClinGen ExAC |
|
| rs960659105 | 41 | E>* | No | Ensembl | |
|
rs960659105 CA257896927 |
41 | E>* | No |
ClinGen Ensembl |
|
|
CA7129567 rs768837042 |
42 | Q>L | No |
ClinGen ExAC |
|
| rs768837042 | 42 | Q>L | No | ExAC | |
| rs1339767718 | 44 | I>N | No | TOPMed | |
|
CA389328132 rs1339767718 |
44 | I>N | No |
ClinGen TOPMed |
|
| rs1393090952 | 45 | Q>R | No | Ensembl | |
|
CA389328124 rs1393090952 |
45 | Q>R | No |
ClinGen Ensembl |
|
|
rs747071344 CA7129566 |
47 | E>K | No |
ClinGen ExAC gnomAD |
|
| rs747071344 | 47 | E>K | No |
ExAC gnomAD |
|
|
CA389328102 rs1275508183 |
48 | L>P | No |
ClinGen gnomAD |
|
| rs1275508183 | 48 | L>P | No | gnomAD | |
|
CA7129565 rs780253706 |
49 | Q>* | No |
ClinGen ExAC gnomAD |
|
| rs780253706 | 49 | Q>* | No |
ExAC gnomAD |
|
|
CA7129564 rs758572495 |
50 | T>I | No |
ClinGen ExAC gnomAD |
|
| rs758572495 | 50 | T>I | No |
ExAC gnomAD |
|
|
rs1291895198 CA389328083 |
51 | A>V | No |
ClinGen Ensembl |
|
| rs1291895198 | 51 | A>V | No | Ensembl | |
| rs779290833 | 52 | K>R | No |
ExAC gnomAD |
|
|
CA7129562 rs779290833 |
52 | K>R | No |
ClinGen ExAC gnomAD |
|
| rs754404734 | 53 | K>N | No |
ExAC gnomAD |
|
|
CA7129561 rs754404734 |
53 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs764525280 CA7129559 |
56 | T>N | No |
ClinGen ExAC gnomAD |
|
| rs764525280 | 56 | T>N | No |
ExAC gnomAD |
|
|
CA7129557 rs752178530 |
58 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs752178530 | 58 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1397169372 | 58 | N>S | No | gnomAD | |
|
rs1397169372 CA389328038 |
58 | N>S | No |
ClinGen gnomAD |
|
| rs941386423 | 60 | R>T | No | TOPMed | |
|
rs941386423 CA257896905 |
60 | R>T | No |
ClinGen TOPMed |
|
|
CA257896850 rs1039775478 |
65 | A>T | No |
ClinGen TOPMed |
|
| rs1039775478 | 65 | A>T | No | TOPMed | |
|
rs1341250015 CA389327982 |
65 | A>V | No |
ClinGen TOPMed |
|
| rs1341250015 | 65 | A>V | No | TOPMed | |
|
CA389327972 rs145042317 |
67 | R>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs145042317 | 67 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
|
rs141054901 CA7129539 |
67 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs141054901 | 67 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145042317 | 67 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
|
CA7129540 rs145042317 |
67 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs370444152 | 69 | K>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs370444152 CA7129538 |
69 | K>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs556369622 | 72 | F>Y | No |
ExAC gnomAD |
|
|
rs556369622 CA7129537 |
72 | F>Y | No |
ClinGen ExAC gnomAD |
|
|
CA7129534 rs751315520 |
73 | E>A | No |
ClinGen ExAC gnomAD |
|
| rs751315520 | 73 | E>A | No |
ExAC gnomAD |
|
|
rs575738905 CA7129535 |
73 | E>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| rs575738905 | 73 | E>K | No |
1000Genomes ExAC gnomAD |
|
|
CA257896818 rs201480938 |
74 | Q>* | No |
ClinGen gnomAD |
|
| rs201480938 | 74 | Q>* | No | gnomAD | |
|
CA7129533 rs766142791 |
74 | Q>H | No |
ClinGen ExAC gnomAD |
|
| rs766142791 | 74 | Q>H | No |
ExAC gnomAD |
|
| rs1210651507 | 75 | Q>L | No | gnomAD | |
|
rs1210651507 CA389327918 |
75 | Q>L | No |
ClinGen gnomAD |
|
| rs1339217657 | 77 | A>P | No |
TOPMed gnomAD |
|
|
rs1339217657 CA389327907 |
77 | A>P | No |
ClinGen TOPMed gnomAD |
|
| rs12436892 | 77 | A>V | No |
ExAC TOPMed gnomAD |
|
|
rs12436892 CA7129532 |
77 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs750214495 | 80 | D>N | No |
ExAC gnomAD |
|
|
CA7129531 rs750214495 |
80 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA7129530 rs374883618 |
81 | G>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs374883618 | 81 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1385850532 | 82 | T>I | No | TOPMed | |
|
rs1385850532 CA389327873 |
82 | T>I | No |
ClinGen TOPMed |
|
| rs772101723 | 82 | T>P | No |
ExAC gnomAD |
|
|
CA7129527 rs772101723 |
82 | T>P | No |
ClinGen ExAC gnomAD |
|
| rs576790627 | 84 | S>A | No |
1000Genomes ExAC gnomAD |
|
|
CA7129526 rs576790627 |
84 | S>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1383821381 CA389327855 |
85 | T>I | No |
ClinGen TOPMed |
|
| rs1383821381 | 85 | T>I | No | TOPMed | |
|
rs771177184 CA7129523 |
90 | R>C | No |
ClinGen ExAC gnomAD |
|
| rs771177184 | 90 | R>C | No |
ExAC gnomAD |
|
| rs1394432466 | 90 | R>H | No |
TOPMed gnomAD |
|
|
CA389327822 rs1394432466 |
90 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs749603690 CA7129522 |
91 | E>* | No |
ClinGen ExAC |
|
| rs749603690 | 91 | E>* | No | ExAC | |
| rs778044294 | 92 | A>S | No |
ExAC TOPMed |
|
|
CA257896766 rs778044294 |
92 | A>S | No |
ClinGen ExAC TOPMed |
|
|
CA7129521 rs778044294 |
92 | A>T | No |
ClinGen ExAC TOPMed |
|
| rs778044294 | 92 | A>T | No |
ExAC TOPMed |
|
|
CA7129520 rs370939325 |
93 | I>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs370939325 | 93 | I>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs748665711 | 94 | E>K | No |
ExAC gnomAD |
|
|
CA7129519 rs748665711 |
94 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs375962047 CA257896755 |
96 | A>D | No |
ClinGen Ensembl |
|
| rs375962047 | 96 | A>D | No | Ensembl | |
| rs1199904250 | 97 | T>A | No |
TOPMed gnomAD |
|
|
rs1199904250 CA389327782 |
97 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
CA257896754 rs780740011 |
97 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs780740011 | 97 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs780740011 | 97 | T>S | No |
ExAC TOPMed gnomAD |
|
|
rs780740011 CA7129518 |
97 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs534511022 | 98 | T>A | No |
1000Genomes ExAC gnomAD |
|
|
CA7129517 rs534511022 |
98 | T>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA389327773 rs1483548351 |
99 | N>D | No |
ClinGen gnomAD |
|
| rs1483548351 | 99 | N>D | No | gnomAD | |
|
rs751117443 CA7129516 |
100 | A>G | No |
ClinGen ExAC gnomAD |
|
| rs751117443 | 100 | A>G | No |
ExAC gnomAD |
|
| rs1280258982 | 100 | A>S | No | gnomAD | |
|
CA389327766 rs1280258982 |
100 | A>S | No |
ClinGen gnomAD |
|
| rs1346592613 | 103 | L>F | No |
TOPMed gnomAD |
|
|
CA389327745 rs1346592613 |
103 | L>F | No |
ClinGen TOPMed gnomAD |
|
| rs145600912 | 104 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7129515 rs145600912 |
104 | R>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs145806351 CA7129513 |
104 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145806351 | 104 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145806351 | 104 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs145806351 CA7129514 |
104 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1410210575 | 106 | M>I | No | gnomAD | |
|
CA389327727 rs1410210575 |
106 | M>I | No |
ClinGen gnomAD |
|
| rs1194654619 | 106 | M>T | No | TOPMed | |
|
CA389327729 rs1194654619 |
106 | M>T | No |
ClinGen TOPMed |
|
| rs555204117 | 106 | M>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
CA7129512 rs555204117 |
106 | M>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs538400003 CA7129510 |
107 | E>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs538400003 | 107 | E>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs141941051 CA7129511 |
107 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs141941051 | 107 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
|
rs141941051 CA389327724 |
107 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs141941051 | 107 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs371123322 CA7129509 |
109 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs371123322 | 109 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1455433984 CA389327707 |
110 | A>T | No |
ClinGen TOPMed |
|
| rs1455433984 | 110 | A>T | No | TOPMed | |
| rs1173258513 | 116 | A>S | No | gnomAD | |
|
rs1173258513 CA389327658 |
116 | A>S | No |
ClinGen gnomAD |
|
| rs1469657762 | 117 | Y>C | No |
TOPMed gnomAD |
|
|
CA389327650 rs1469657762 |
117 | Y>C | No |
ClinGen TOPMed gnomAD |
|
| rs373027314 | 118 | Q>* | No |
ExAC TOPMed gnomAD |
|
|
rs373027314 CA7129508 |
118 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs373027314 CA389327645 |
118 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs373027314 | 118 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs1184543755 | 119 | D>H | No | gnomAD | |
|
rs1184543755 CA389327638 |
119 | D>H | No |
ClinGen gnomAD |
|
| rs766509120 | 120 | M>I | No |
ExAC gnomAD |
|
|
CA7129486 rs766509120 |
120 | M>I | No |
ClinGen ExAC gnomAD |
|
| rs373464425 | 122 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
|
rs373464425 CA7129485 |
122 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs199498643 | 123 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
|
rs199498643 CA7129484 |
123 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA257896435 rs199498643 |
123 | D>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs199498643 | 123 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
|
CA389327581 rs1162859213 |
124 | K>R | No |
ClinGen gnomAD |
|
| rs1162859213 | 124 | K>R | No | gnomAD | |
| rs1391850402 | 126 | D>V | No | TOPMed | |
|
rs1391850402 CA389327554 |
126 | D>V | No |
ClinGen TOPMed |
|
| rs147600552 | 129 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
|
rs147600552 CA7129481 |
129 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs762042465 | 129 | M>R | No |
ExAC TOPMed gnomAD |
|
|
CA7129483 rs762042465 |
129 | M>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs762042465 | 129 | M>T | No |
ExAC TOPMed gnomAD |
|
|
rs762042465 CA7129482 |
129 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs769256837 | 132 | I>T | No |
ExAC gnomAD |
|
|
rs769256837 CA7129480 |
132 | I>T | No |
ClinGen ExAC gnomAD |
|
| rs1448186476 | 132 | I>V | No | gnomAD | |
|
CA389327488 rs1448186476 |
132 | I>V | No |
ClinGen gnomAD |
|
|
CA7129479 rs145119541 |
133 | T>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs145119541 | 133 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs771651000 | 134 | E>G | No |
ExAC gnomAD |
|
|
rs771651000 CA7129477 |
134 | E>G | No |
ClinGen ExAC gnomAD |
|
| rs117552046 | 136 | Q>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs117552046 CA7129476 |
136 | Q>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7129475 rs778393755 |
136 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs778393755 | 136 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs749017330 | 141 | Q>H | No |
ExAC gnomAD |
|
|
CA7129473 rs749017330 |
141 | Q>H | No |
ClinGen ExAC gnomAD |
|
| rs1348752985 | 141 | Q>R | No | TOPMed | |
|
rs1348752985 CA389327373 |
141 | Q>R | No |
ClinGen TOPMed |
|
|
rs777582416 CA7129472 |
142 | I>M | No |
ClinGen ExAC gnomAD |
|
| rs777582416 | 142 | I>M | No |
ExAC gnomAD |
|
|
CA389327333 rs371942693 |
144 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs371942693 | 144 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
|
rs375056533 CA7129471 |
144 | D>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs375056533 | 144 | D>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs375056533 | 144 | D>V | No |
ESP ExAC TOPMed gnomAD |
|
|
CA7129470 rs375056533 |
144 | D>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs151089933 CA7129467 |
148 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs151089933 | 148 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs758343224 | 148 | R>W | No |
ExAC TOPMed gnomAD |
|
|
CA7129468 rs758343224 |
148 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs1438029866 | 150 | M>K | No | TOPMed | |
|
CA389327298 rs1438029866 |
150 | M>K | No |
ClinGen TOPMed |
|
| rs371342323 | 150 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs371342323 CA7129466 |
150 | M>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7129465 rs371342323 |
150 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs371342323 | 150 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs754252657 | 151 | G>V | No |
ExAC gnomAD |
|
|
CA7129464 rs754252657 |
151 | G>V | No |
ClinGen ExAC gnomAD |
|
|
VAR_023384 CA7129462 rs2295322 |
153 | G>R | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2295322 | 153 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1594469418 | 154 | D>N | No | Ensembl | |
|
CA389327275 rs1594469418 |
154 | D>N | No |
ClinGen Ensembl |
|
| rs1169721543 | 155 | D>Y | No | gnomAD | |
|
rs1169721543 CA389327268 |
155 | D>Y | No |
ClinGen gnomAD |
|
| rs757409020 | 160 | E>A | No |
ExAC TOPMed gnomAD |
|
|
CA7129444 rs757409020 |
160 | E>A | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs754056488 | 161 | L>R | No |
ExAC gnomAD |
|
|
rs754056488 CA7129443 |
161 | L>R | No |
ClinGen ExAC gnomAD |
|
| rs761084506 | 168 | L>P | No |
ExAC TOPMed gnomAD |
|
|
rs761084506 CA7129441 |
168 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764417489 CA7129442 |
168 | L>V | No |
ClinGen ExAC gnomAD |
|
| rs764417489 | 168 | L>V | No |
ExAC gnomAD |
|
|
rs753138859 CA7129439 |
169 | E>K | No |
ClinGen ExAC gnomAD |
|
| rs753138859 | 169 | E>K | No |
ExAC gnomAD |
|
|
CA389327137 rs1288026756 |
172 | E>K | No |
ClinGen gnomAD |
|
| rs1288026756 | 172 | E>K | No | gnomAD | |
| rs760132790 | 173 | L>M | No |
ExAC gnomAD |
|
|
CA7129437 rs760132790 |
173 | L>M | No |
ClinGen ExAC gnomAD |
|
| rs773725812 | 174 | A>T | No |
ExAC gnomAD |
|
|
CA7129436 rs773725812 |
174 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA7129435 rs148589696 |
176 | E>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs148589696 | 176 | E>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7129433 rs772780946 |
179 | N>K | No |
ClinGen ExAC gnomAD |
|
| rs772780946 | 179 | N>K | No |
ExAC gnomAD |
|
|
rs762459227 CA7129434 |
179 | N>S | No |
ClinGen ExAC gnomAD |
|
| rs762459227 | 179 | N>S | No |
ExAC gnomAD |
|
| rs1368954055 | 180 | V>A | No | TOPMed | |
|
CA389327081 rs1368954055 |
180 | V>A | No |
ClinGen TOPMed |
|
|
rs1305039283 CA389327083 |
180 | V>M | No |
ClinGen TOPMed gnomAD |
|
| rs1305039283 | 180 | V>M | No |
TOPMed gnomAD |
|
|
CA257896163 rs1037307556 |
182 | D>N | No |
ClinGen gnomAD |
|
| rs1037307556 | 182 | D>N | No | gnomAD | |
|
rs747889785 CA7129431 |
184 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs747889785 | 184 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1458457056 | 185 | E>A | No |
TOPMed gnomAD |
|
|
CA389327047 rs1458457056 |
185 | E>A | No |
ClinGen TOPMed gnomAD |
|
| rs146389065 | 186 | E>Q | No | ESP | |
|
CA257896155 rs146389065 |
186 | E>Q | No |
ClinGen ESP |
|
| rs781087736 | 187 | P>A | No |
ExAC TOPMed |
|
|
rs781087736 CA7129430 |
187 | P>A | No |
ClinGen ExAC TOPMed |
|
|
rs768445146 CA7129428 |
190 | K>E | No |
ClinGen ExAC gnomAD |
|
| rs768445146 | 190 | K>E | No |
ExAC gnomAD |
|
| rs1414990338 | 195 | P>S | No | gnomAD | |
|
rs1414990338 CA389326985 |
195 | P>S | No |
ClinGen gnomAD |
|
| rs1258201720 | 197 | T>A | No | gnomAD | |
|
CA389326972 rs1258201720 |
197 | T>A | No |
ClinGen gnomAD |
|
| rs76444629 | 198 | H>Y | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7129425 rs76444629 |
198 | H>Y | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs910438479 CA257896117 |
199 | L>P | No |
ClinGen Ensembl |
|
| rs910438479 | 199 | L>P | No | Ensembl | |
|
CA7129424 rs377089196 |
200 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs377089196 | 200 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
|
CA389326948 rs1331512011 |
201 | A>V | No |
ClinGen gnomAD |
|
| rs1331512011 | 201 | A>V | No | gnomAD | |
|
rs1268956735 CA389326944 |
202 | G>R | No |
ClinGen gnomAD |
|
| rs1268956735 | 202 | G>R | No | gnomAD | |
|
CA7129407 rs749360328 |
204 | A>G | No |
ClinGen ExAC gnomAD |
|
| rs749360328 | 204 | A>G | No |
ExAC gnomAD |
|
|
CA389326933 rs1594469195 |
204 | A>P | No |
ClinGen Ensembl |
|
| rs1594469195 | 204 | A>P | No | Ensembl | |
|
CA7129408 rs749360328 |
204 | A>V | No |
ClinGen ExAC gnomAD |
|
| rs749360328 | 204 | A>V | No |
ExAC gnomAD |
|
|
CA7129406 rs143736280 |
205 | P>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs143736280 | 205 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1353153654 | 206 | K>E | No | gnomAD | |
|
rs1353153654 CA389325939 |
206 | K>E | No |
ClinGen gnomAD |
|
| rs756403727 | 206 | K>N | No |
ExAC gnomAD |
|
|
rs756403727 CA7129405 |
206 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1322556245 CA389325891 |
208 | D>G | No |
ClinGen gnomAD |
|
| rs1322556245 | 208 | D>G | No | gnomAD | |
|
rs748457977 CA7129404 |
208 | D>H | No |
ClinGen ExAC gnomAD |
|
| rs748457977 | 208 | D>H | No |
ExAC gnomAD |
|
| rs371017683 | 209 | E>K | No | Ensembl | |
|
CA257895883 rs371017683 |
209 | E>K | No |
ClinGen Ensembl |
|
| rs1384686477 | 210 | D>G | No | gnomAD | |
|
rs1384686477 CA389325847 |
210 | D>G | No |
ClinGen gnomAD |
|
|
CA389325761 rs1392681413 |
213 | A>V | No |
ClinGen TOPMed gnomAD |
|
| rs1392681413 | 213 | A>V | No |
TOPMed gnomAD |
|
|
rs1293919805 CA389325692 |
218 | A>P | No |
ClinGen gnomAD |
|
| rs1293919805 | 218 | A>P | No | gnomAD | |
| rs537771236 | 218 | A>V | No |
1000Genomes ExAC gnomAD |
|
|
CA7129402 rs537771236 |
218 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs755299003 CA7129400 |
220 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA389325637 rs1390856419 |
220 | W>* | No |
ClinGen gnomAD |
|
| rs1390856419 | 220 | W>* | No | gnomAD | |
| rs755299003 | 220 | W>* | No |
ExAC gnomAD |
|
|
CA389325638 rs1390856419 |
220 | W>C | No |
ClinGen gnomAD |
|
| rs1390856419 | 220 | W>C | No | gnomAD | |
|
rs1007849983 CA257895873 |
222 | S>P | No |
ClinGen TOPMed |
|
| rs1007849983 | 222 | S>P | No | TOPMed | |
|
rs1198467833 CA389325585 |
223 | S>W | No |
ClinGen gnomAD |
|
| rs1198467833 | 223 | S>W | No | gnomAD |
No associated diseases with Q9BY43
No regional properties for Q9BY43
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q9BY43 | |||
Functions
16 GO annotations of cellular component
| Name | Definition |
|---|---|
| amphisome membrane | Any membrane that is part of an amphisome. |
| autophagosome membrane | The lipid bilayer surrounding an autophagosome, a double-membrane-bounded vesicle in which endogenous cellular material is sequestered. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic side of plasma membrane | The leaflet the plasma membrane that faces the cytoplasm and any proteins embedded or anchored in it or attached to its surface. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| ESCRT III complex | A complex with membrane scission activity that plays a major role in many processes where membranes are remodelled - including endosomal transport (vesicle budding), nuclear envelope organisation (membrane closure, mitotic bridge cleavage), and cytokinesis (abscission). |
| kinetochore | A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules. |
| kinetochore microtubule | Any of the spindle microtubules that attach to the kinetochores of chromosomes by their plus ends, and maneuver the chromosomes during mitotic or meiotic chromosome segregation. |
| lysosomal membrane | The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm. |
| membrane coat | Any of several different proteinaceous coats that can associate with membranes. Membrane coats include those formed by clathrin plus an adaptor complex, the COPI and COPII complexes, and possibly others. They are found associated with membranes on many vesicles as well as other membrane features such as pits and perhaps tubules. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| multivesicular body | A type of endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body membrane | The lipid bilayer surrounding a multivesicular body. |
| nuclear pore | A protein complex providing a discrete opening in the nuclear envelope of a eukaryotic cell, where the inner and outer nuclear membranes are joined. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATPase binding | Binding to an ATPase, any enzyme that catalyzes the hydrolysis of ATP. |
| identical protein binding | Binding to an identical protein or proteins. |
| lipid binding | Binding to a lipid. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
24 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome maturation | Removal of PI3P and Atg8/LC3 after the closure of the phagophore and before the fusion with the endosome/lysosome (e.g. mammals and insects) or vacuole (yeast), and that very likely destabilizes other Atg proteins and thus enables their efficient dissociation and recycling. |
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| late endosome to lysosome transport | The directed movement of substances from late endosome to lysosome. |
| late endosome to vacuole transport via multivesicular body sorting pathway | The directed movement of substances from endosomes to vacuoles by a pathway in which molecules are sorted into multivesicular bodies, which then fuse with the vacuole. |
| macroautophagy | The major inducible pathway for the general turnover of cytoplasmic constituents in eukaryotic cells, it is also responsible for the degradation of active cytoplasmic enzymes and organelles during nutrient starvation. Macroautophagy involves the formation of double-membrane-bounded autophagosomes which enclose the cytoplasmic constituent targeted for degradation in a membrane-bounded structure. Autophagosomes then fuse with a lysosome (or vacuole) releasing single-membrane-bounded autophagic bodies that are then degraded within the lysosome (or vacuole). Some types of macroautophagy, e.g. pexophagy, mitophagy, involve selective targeting of the targets to be degraded. |
| membrane invagination | The infolding of a membrane. |
| midbody abscission | The process by which the midbody, the cytoplasmic bridge that connects the two prospective daughter cells, is severed at the end of mitotic cytokinesis, resulting in two separate daughter cells. |
| mitotic metaphase plate congression | The cell cycle process in which chromosomes are aligned at the metaphase plate, a plane halfway between the poles of the mitotic spindle, during mitosis. |
| multivesicular body assembly | The aggregation, arrangement and bonding together of a set of components to form a multivesicular body, a type of late endosome in which regions of the limiting endosomal membrane invaginate to form internal vesicles; membrane proteins that enter the internal vesicles are sequestered from the cytoplasm. |
| multivesicular body sorting pathway | A vesicle-mediated transport process in which transmembrane proteins are ubiquitylated to facilitate their entry into luminal vesicles of multivesicular bodies (MVBs); upon subsequent fusion of MVBs with lysosomes or vacuoles, the cargo proteins are degraded. |
| negative regulation of autophagosome assembly | Any process that stops, prevents or reduces the frequency, rate or extent of autophagosome assembly. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| negative regulation of neuron death | Any process that stops, prevents or reduces the frequency, rate or extent of neuron death. |
| nuclear membrane reassembly | The reformation of the nuclear membranes following their breakdown in the context of a normal process. |
| nucleus organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the nucleus. |
| plasma membrane repair | The resealing of a cell plasma membrane after cellular wounding due to, for instance, mechanical stress. |
| plasma membrane tubulation | A membrane tubulation process occurring in a plasma membrane. |
| posttranslational protein targeting to endoplasmic reticulum membrane | The targeting of proteins to a membrane that occurs after their translation. Some secretory proteins exhibit posttranslational transport into the endoplasmic reticulum (ER) lumen: they are synthesized in their entirety on free cytosolic ribosomes and then released into the cytosol, where they are bound by chaperones which keep them in an unfolded state, and subsequently are translocated across the ER membrane. |
| protein polymerization | The process of creating protein polymers, compounds composed of a large number of component monomers; polymeric proteins may be made up of different or identical monomers. Polymerization occurs by the addition of extra monomers to an existing poly- or oligomeric protein. |
| regulation of mitotic spindle assembly | Any process that modulates the frequency, rate or extent of mitotic spindle assembly. |
| ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | The chemical reactions and pathways resulting in the breakdown of a protein or peptide covalently tagged with ubiquitin, via the multivesicular body (MVB) sorting pathway; ubiquitin-tagged proteins are sorted into MVBs, and delivered to a lysosome/vacuole for degradation. |
| vesicle budding from membrane | The evagination of a membrane, resulting in formation of a vesicle. |
| viral budding from plasma membrane | A viral budding that starts with formation of a membrane curvature in the host plasma membrane. |
| viral budding via host ESCRT complex | Viral budding which uses a host ESCRT protein complex, or complexes, to mediate the budding process. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q96FZ7 | CHMP6 | Charged multivesicular body protein 6 | Homo sapiens (Human) | SS |
| Q9D7F7 | Chmp4c | Charged multivesicular body protein 4c | Mus musculus (Mouse) | SS |
| Q9D8B3 | Chmp4b | Charged multivesicular body protein 4b | Mus musculus (Mouse) | SS |
| Q569C1 | Chmp4c | Charged multivesicular body protein 4c | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSGLGRLFGK | GKKEKGPTPE | EAIQKLKETE | KILIKKQEFL | EQKIQQELQT | AKKYGTKNKR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AALQALRRKK | RFEQQLAQTD | GTLSTLEFQR | EAIENATTNA | EVLRTMELAA | QSMKKAYQDM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DIDKVDELMT | DITEQQEVAQ | QISDAISRPM | GFGDDVDEDE | LLEELEELEQ | EELAQELLNV |
| 190 | 200 | 210 | 220 | ||
| GDKEEEPSVK | LPSVPSTHLP | AGPAPKVDED | EEALKQLAEW | VS |