Q9BWU1
Gene name |
CDK19 (CDC2L6, CDK11, KIAA1028) |
Protein name |
Cyclin-dependent kinase 19 |
Names |
CDC2-related protein kinase 6, Cell division cycle 2-like protein kinase 6, Cell division protein kinase 19, Cyclin-dependent kinase 11, Death-preventing kinase |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:23097 |
EC number |
2.7.11.22: Protein-serine/threonine kinases |
Protein Class |
|
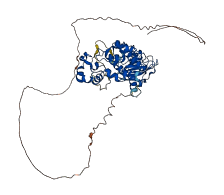
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
172-198 (Activation loop from InterPro)
Target domain |
21-335 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q9BWU1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9BWU1-F1 | Predicted | AlphaFoldDB |
225 variants for Q9BWU1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1783518890 RCV001250468 VAR_084392 |
28 | G>A | DEE87 [UniProt] | Yes |
ClinVar UniProt dbSNP |
|
RCV001252679 RCV001250465 rs1783519120 VAR_084393 RCV001252678 |
28 | G>R | Developmental and epileptic encephalopathy, 87 DEE87; decreased protein kinase activity [ClinVar, UniProt] | Yes |
ClinVar dbSNP UniProt |
|
rs1783518295 RCV001250507 |
31 | T>N | Developmental and epileptic encephalopathy, 87 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1783517622 RCV001252680 VAR_084394 |
32 | Y>C | Developmental and epileptic encephalopathy, 87 DEE87 [ClinVar, UniProt] | Yes |
ClinVar UniProt dbSNP |
|
RCV001195090 rs1236246272 CA365245579 VAR_084395 |
32 | Y>H | Developmental and epileptic encephalopathy, 87 DEE87; fails to rescue neurologic phenotypes in a Drosophila model system; increased protein kinase activity [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt dbSNP gnomAD |
|
rs1779473650 RCV001255605 RCV002560185 TCGA novel RCV001195089 VAR_084396 |
196 | T>A | Developmental and epileptic encephalopathy, 87 Variant assessed as Somatic; impact. Inborn genetic diseases DEE87; fails to rescue neurologic phenotypes in a Drosophila model system [ClinVar, NCI-TCGA, UniProt] | Yes |
ClinVar NCI-TCGA UniProt dbSNP |
|
rs1779473436 VAR_084397 RCV001252681 |
197 | F>L | Developmental and epileptic encephalopathy, 87 DEE87 [ClinVar, UniProt] | Yes |
ClinVar UniProt dbSNP |
|
rs1779473213 VAR_084398 RCV001252682 |
198 | W>C | Developmental and epileptic encephalopathy, 87 DEE87 [ClinVar, UniProt] | Yes |
ClinVar UniProt dbSNP |
|
VAR_084399 rs1779472995 RCV001250464 |
200 | R>W | DEE87 [UniProt] | Yes |
ClinVar UniProt dbSNP |
|
RCV001252732 CA3958895 rs55914301 COSM39303 |
395 | A>V | central_nervous_system Microcephaly [Cosmic, ClinVar] | Yes |
ClinGen cosmic curated ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
| TCGA novel | 3 | Y>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365245767 rs1404396903 |
4 | D>G | No |
ClinGen gnomAD |
|
|
CA365245727 rs1562311725 |
10 | A>T | No |
ClinGen Ensembl |
|
|
rs757584020 CA3959241 |
12 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs145535321 CA145220410 |
13 | R>W | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1156998824 CA365245705 |
14 | E>K | No |
ClinGen gnomAD |
|
|
rs760961719 CA3959238 |
18 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA3959239 rs764169323 |
18 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA365245599 rs1483542851 |
29 | R>S | No |
ClinGen gnomAD |
|
|
rs1324013746 CA365245550 |
36 | Y>C | No |
ClinGen TOPMed |
|
|
CA365245541 rs1241915940 |
37 | K>R | No |
ClinGen gnomAD |
|
|
CA3959234 rs773263211 |
40 | R>G | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs773263211 CA365245524 |
40 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs761477193 CA3959232 |
43 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 44 | K>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1462361150 CA365355594 |
45 | D>E | No |
ClinGen TOPMed |
|
|
rs1385019803 CA365355600 |
45 | D>N | No |
ClinGen gnomAD |
|
|
CA365355597 rs1290014343 |
45 | D>V | No |
ClinGen gnomAD |
|
|
CA3959168 rs762489169 |
48 | E>K | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 57 | T>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3959165 rs748670469 |
59 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365355499 rs748670469 |
59 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777345140 CA3959164 |
60 | S>A | No |
ClinGen ExAC |
|
|
rs1487705565 CA365355410 |
65 | R>T | No |
ClinGen gnomAD |
|
|
CA3959120 rs759991863 |
71 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA365354321 rs1297296367 |
77 | N>T | No |
ClinGen TOPMed |
|
| TCGA novel | 78 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1229122867 CA365354287 |
82 | Q>R | No |
ClinGen gnomAD |
|
|
CA365354279 rs1334230328 |
83 | K>R | No |
ClinGen gnomAD |
|
|
CA365354247 COSM1634423 rs1582824930 |
88 | H>Y | liver [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA3959112 rs758096037 |
96 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs750261087 CA3959111 |
100 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1219070923 CA365354057 COSM201168 |
112 | R>C | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 113 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365354024 rs1426767970 |
117 | N>S | No |
ClinGen gnomAD |
|
|
CA365353994 rs1317130024 |
121 | M>R | No |
ClinGen TOPMed |
|
|
rs1275907568 CA365353996 |
121 | M>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1233609881 CA365353981 |
122 | Q>H | No |
ClinGen TOPMed |
|
|
CA145825554 rs980853034 |
124 | P>S | No |
ClinGen gnomAD |
|
|
CA365353973 rs980853034 |
124 | P>T | No |
ClinGen gnomAD |
|
|
CA365353968 rs1290455860 |
125 | R>G | No |
ClinGen gnomAD |
|
|
CA145825553 rs554919650 |
127 | M>T | No |
ClinGen Ensembl |
|
|
rs748931875 CA3959084 |
128 | V>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA145825552 rs200697462 |
128 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA3959085 rs200697462 |
128 | V>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1209146999 CA365353921 |
132 | L>F | No |
ClinGen TOPMed |
|
|
rs1259552312 CA365353861 |
140 | H>Q | No |
ClinGen gnomAD |
|
|
CA3959081 rs752285573 |
143 | H>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 145 | N>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 152 | L>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs758734214 CA3959059 |
160 | M>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758734214 CA3959060 |
160 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3959058 rs750806739 |
161 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs765317905 CA3959057 |
165 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs762098741 CA3959056 |
168 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA365357695 rs1214040281 |
169 | V>D | No |
ClinGen gnomAD |
|
| TCGA novel | 176 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365357602 rs1386634373 |
181 | N>H | No |
ClinGen TOPMed |
|
|
CA365357598 rs1562139375 |
181 | N>S | No |
ClinGen Ensembl |
|
| TCGA novel | 182 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1325061809 CA365357583 |
183 | P>L | No |
ClinGen gnomAD |
|
| TCGA novel | 183 | P>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1562139316 CA365357518 |
193 | V>G | No |
ClinGen Ensembl |
|
| TCGA novel | 202 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA145821935 rs940533589 |
203 | E>K | No |
ClinGen Ensembl |
|
|
CA365357421 rs1193696403 |
208 | A>G | No |
ClinGen gnomAD |
|
|
CA365357417 rs1373241481 |
209 | R>K | No |
ClinGen gnomAD |
|
|
CA145821431 rs916860957 |
218 | W>* | No |
ClinGen TOPMed |
|
|
rs1055319082 CA145821430 |
228 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs953895657 COSM174895 CA145821429 |
230 | S>L | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
| TCGA novel | 235 | H>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3959018 rs749643967 |
237 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs756420255 CA3959016 |
241 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA145821428 rs780608956 |
241 | I>V | No |
ClinGen Ensembl |
|
|
CA365357150 rs1275224408 |
245 | N>Y | No |
ClinGen gnomAD |
|
|
CA145821426 rs368779010 |
249 | H>N | No |
ClinGen ExAC gnomAD |
|
|
rs368779010 CA3959015 |
249 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1349823433 CA365357087 COSM260037 |
254 | R>W | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA365357039 rs1306900740 |
260 | G>E | No |
ClinGen gnomAD |
|
|
rs865823701 CA145821424 |
263 | A>V | No |
ClinGen Ensembl |
|
|
CA145821414 rs1055234651 |
264 | D>G | No |
ClinGen Ensembl |
|
| TCGA novel | 271 | R>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs865965891 CA145821413 |
273 | M>K | No |
ClinGen Ensembl |
|
|
CA3958991 rs751624560 |
273 | M>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 277 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1410735559 CA365356908 |
277 | P>T | No |
ClinGen gnomAD |
|
|
rs187493296 CA3958989 |
287 | T>A | No |
ClinGen 1000Genomes ExAC |
|
|
rs753876969 CA3958988 |
287 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs1273781226 CA365356797 |
291 | S>N | No |
ClinGen gnomAD |
|
|
CA3958968 rs753749481 |
294 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs200880669 CA3958969 |
294 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1373271422 CA365356755 |
297 | M>K | No |
ClinGen TOPMed |
|
|
rs777727767 CA3958967 |
297 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs1363705470 CA365356731 |
300 | H>Y | No |
ClinGen gnomAD |
|
|
rs767097773 CA3958964 |
301 | K>N | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 304 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1433795864 CA365356633 |
312 | L>F | No |
ClinGen gnomAD |
|
|
CA3958945 rs754862400 |
314 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365356599 rs762517170 |
317 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs762517170 CA3958942 |
317 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA145821012 rs921734283 |
324 | I>N | No |
ClinGen TOPMed |
|
|
CA365356540 rs1481343583 |
326 | S>A | No |
ClinGen gnomAD |
|
|
rs975083127 CA145821011 |
326 | S>L | No |
ClinGen TOPMed |
|
|
rs761138528 CA3958937 |
333 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA365356493 rs1280052591 |
333 | P>H | No |
ClinGen TOPMed |
|
|
rs761138528 CA3958936 |
333 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1282897871 CA365356486 |
334 | Y>C | No |
ClinGen gnomAD |
|
| TCGA novel | 334 | Y>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365356471 rs1237424581 |
336 | Q>R | No |
ClinGen gnomAD |
|
|
CA365356458 rs1316813423 |
338 | D>N | No |
ClinGen gnomAD |
|
|
rs1353016152 CA365356449 |
339 | P>A | No |
ClinGen TOPMed |
|
|
rs1244482015 CA365356380 |
347 | A>V | No |
ClinGen TOPMed |
|
|
CA3958917 rs760899404 |
348 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1293726339 CA365356355 |
351 | I>V | No |
ClinGen gnomAD |
|
|
CA365356329 rs1380275260 |
354 | P>L | No |
ClinGen gnomAD |
|
|
CA365356327 rs1383983955 |
355 | K>E | No |
ClinGen gnomAD |
|
|
CA365356319 rs1289319819 |
356 | R>* | No |
ClinGen gnomAD |
|
|
rs1239138510 CA365356316 |
356 | R>Q | No |
ClinGen TOPMed |
|
|
CA365356301 rs1455274232 |
358 | F>L | No |
ClinGen gnomAD |
|
| TCGA novel | 359 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1261907348 CA365356281 |
361 | E>G | No |
ClinGen TOPMed |
|
|
CA365356261 rs1392285893 |
364 | P>T | No |
ClinGen gnomAD |
|
|
rs375353769 CA145820967 |
368 | G>R | No |
ClinGen ESP |
|
| TCGA novel | 369 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1251968288 CA365356183 |
372 | Q>H | No |
ClinGen TOPMed |
|
|
rs756783771 CA3958899 |
377 | N>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764451268 CA3958897 |
379 | H>Y | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 381 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1252871484 CA365356119 |
381 | Q>E | No |
ClinGen gnomAD |
|
|
CA365356113 rs1179651343 |
381 | Q>H | No |
ClinGen TOPMed |
|
|
rs1410791361 CA365356109 |
382 | P>L | No |
ClinGen TOPMed |
|
|
CA365356101 rs1194812139 |
383 | T>I | No |
ClinGen gnomAD |
|
|
rs1469838359 CA365356092 |
385 | P>S | No |
ClinGen gnomAD |
|
|
CA365356072 rs1248495183 |
388 | Q>E | No |
ClinGen gnomAD |
|
|
rs1195371419 CA365356071 |
388 | Q>P | No |
ClinGen gnomAD |
|
|
CA3958896 rs756647858 |
389 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA365356056 rs1253352940 |
390 | A>V | No |
ClinGen gnomAD |
|
|
rs1174138651 CA365356027 |
395 | A>T | No |
ClinGen TOPMed |
|
|
CA3958893 rs759944605 |
396 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1329446147 CA365356018 |
397 | P>A | No |
ClinGen TOPMed |
|
|
CA145820913 rs546919397 |
397 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs546919397 CA3958892 |
397 | P>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1582691168 CA365356013 |
398 | P>S | No |
ClinGen Ensembl |
|
| TCGA novel | 400 | Q>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3958891 rs766619789 |
402 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA3958890 rs762896900 |
403 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA365355953 rs1309504394 |
406 | N>K | No |
ClinGen TOPMed gnomAD |
|
|
rs773360002 CA3958889 |
407 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA365355946 rs1394872857 |
408 | T>P | No |
ClinGen gnomAD |
|
|
rs570611641 CA3958887 |
409 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365355936 rs1272012303 |
409 | A>V | No |
ClinGen gnomAD |
|
|
rs1362613045 CA365355932 |
410 | G>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 411 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 412 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365355914 rs1196942838 |
413 | G>R | No |
ClinGen TOPMed |
|
|
rs745684988 CA3958884 |
415 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA3958883 rs548129906 |
416 | V>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA365355893 rs1340287712 COSM1072144 |
417 | G>R | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
CA365355885 rs1562126547 |
418 | G>S | No |
ClinGen Ensembl |
|
|
rs1347007822 CA365355876 |
419 | T>I | No |
ClinGen gnomAD |
|
|
CA3958878 rs145745297 |
420 | G>R | No |
ClinGen ESP ExAC gnomAD |
|
|
rs199931376 CA3958877 COSM1072143 |
421 | A>T | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs1309411704 CA365355866 |
421 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1582690742 CA365355862 |
422 | G>A | No |
ClinGen Ensembl |
|
|
rs911298280 CA145820910 |
423 | L>M | No |
ClinGen Ensembl |
|
|
CA3958876 rs375258247 |
424 | Q>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA365355842 rs1374549254 |
425 | H>R | No |
ClinGen gnomAD |
|
|
CA3958875 rs751944182 |
426 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs766533848 CA3958874 |
428 | D>V | No |
ClinGen ExAC gnomAD |
|
|
CA3958873 rs531311578 |
429 | S>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA3958872 rs750680904 |
430 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA3958871 rs765349281 |
430 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs1172697598 CA365355791 |
433 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
rs761836116 CA3958870 |
434 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs376573708 CA3958868 |
436 | P>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs759343831 CA3958867 |
438 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA365355744 rs1443955357 |
440 | P>A | No |
ClinGen gnomAD |
|
|
CA3958866 rs774291402 |
441 | R>Q | Variant assessed as Somatic; 4.623e-05 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA365355738 rs1281979042 |
441 | R>W | No |
ClinGen gnomAD |
|
|
CA365355719 rs1354410644 |
444 | P>L | No |
ClinGen gnomAD |
|
|
CA365355704 rs1450585259 |
447 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA365355706 rs1450585259 |
447 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA3958862 rs769319056 |
453 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3958861 rs747643636 |
454 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1228619572 CA365355653 |
455 | P>S | No |
ClinGen TOPMed |
|
|
rs781628403 CA3958860 |
456 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1449876565 CA365354912 |
460 | H>R | No |
ClinGen gnomAD |
|
|
CA3958841 rs768226674 |
460 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1191674190 CA365354896 |
461 | S>P | No |
ClinGen gnomAD |
|
|
CA365354875 rs1467593689 |
462 | S>T | No |
ClinGen gnomAD |
|
|
CA3958839 rs373933843 COSM1072141 |
464 | R>C | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs758747416 CA3958838 |
464 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1305376207 CA365354832 |
465 | L>P | No |
ClinGen gnomAD |
|
|
rs779134930 CA3958836 |
470 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764256188 CA3958834 |
471 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764256188 CA3958833 |
471 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA3958832 rs756309724 |
472 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1471050497 CA365354684 |
473 | G>R | No |
ClinGen TOPMed |
|
|
CA365354642 rs1440180529 |
476 | Q>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA145820260 rs1053526679 |
476 | Q>R | No |
ClinGen TOPMed |
|
|
rs1281492474 CA365354582 |
479 | S>C | No |
ClinGen gnomAD |
|
|
rs1582669652 CA365354513 |
485 | S>F | No |
ClinGen Ensembl |
|
|
rs766259977 CA3958829 |
486 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA365354475 rs1309316274 |
491 | S>A | No |
ClinGen TOPMed |
|
|
rs1582669576 CA365354452 |
494 | H>P | No |
ClinGen Ensembl |
|
|
CA145820257 rs868855764 |
495 | P>L | No |
ClinGen Ensembl |
|
|
CA145820258 rs928696407 |
495 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA365354425 rs1171590583 |
498 | Q>R | No |
ClinGen gnomAD |
|
|
rs1562119282 CA365354416 |
499 | A>G | No |
ClinGen Ensembl |
|
|
rs1326125252 CA365354421 |
499 | A>T | No |
ClinGen TOPMed |
|
|
rs1310668301 CA365354412 |
500 | H>R | No |
ClinGen gnomAD |
|
|
CA3958824 rs776325246 |
501 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs761454481 CA3958825 |
501 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1195851303 CA365354401 |
502 | Y>C | No |
ClinGen TOPMed |
|
|
CA3958822 rs377400438 |
503 | Y>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
1 associated diseases with Q9BWU1
[MIM: 618916]: Developmental and epileptic encephalopathy 87 (DEE87)
A form of epileptic encephalopathy, a heterogeneous group of severe early-onset epilepsies characterized by refractory seizures, neurodevelopmental impairment, and poor prognosis. Development is normal prior to seizure onset, after which cognitive and motor delays become apparent. DEE87 inheritance is autosomal dominant. {ECO:0000269|PubMed:32330417, ECO:0000269|PubMed:33134521, ECO:0000269|PubMed:33495529}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A form of epileptic encephalopathy, a heterogeneous group of severe early-onset epilepsies characterized by refractory seizures, neurodevelopmental impairment, and poor prognosis. Development is normal prior to seizure onset, after which cognitive and motor delays become apparent. DEE87 inheritance is autosomal dominant. {ECO:0000269|PubMed:32330417, ECO:0000269|PubMed:33134521, ECO:0000269|PubMed:33495529}. Note=The disease is caused by variants affecting the gene represented in this entry.
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.22 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| mediator complex | A protein complex that interacts with the carboxy-terminal domain of the largest subunit of RNA polymerase II and plays an active role in transducing the signal from a transcription factor to the transcriptional machinery. The mediator complex is required for activation of transcription of most protein-coding genes, but can also act as a transcriptional corepressor. The Saccharomyces complex contains several identifiable subcomplexes: a head domain comprising Srb2, -4, and -5, Med6, -8, and -11, and Rox3 proteins; a middle domain comprising Med1, -4, and -7, Nut1 and -2, Cse2, Rgr1, Soh1, and Srb7 proteins; a tail consisting of Gal11p, Med2p, Pgd1p, and Sin4p; and a regulatory subcomplex comprising Ssn2, -3, and -8, and Srb8 proteins. Metazoan mediator complexes have similar modular structures and include homologs of yeast Srb and Med proteins. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| cyclin-dependent protein serine/threonine kinase activity | Cyclin-dependent catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| RNA polymerase II CTD heptapeptide repeat kinase activity | Catalysis of the reaction: ATP + RNA polymerase II large subunit CTD heptapeptide repeat (YSPTSPS) = ADP + H+ + phosphorylated RNA polymerase II. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to lipopolysaccharide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of inflammatory response | Any process that activates or increases the frequency, rate or extent of the inflammatory response. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
19 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P39073 | SSN3 | Meiotic mRNA stability protein kinase SSN3 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q9VT57 | Cdk8 | Cyclin-dependent kinase 8 | Drosophila melanogaster (Fruit fly) | PR |
| Q15131 | CDK10 | Cyclin-dependent kinase 10 | Homo sapiens (Human) | PR |
| Q9UQ88 | CDK11A | Cyclin-dependent kinase 11A | Homo sapiens (Human) | PR |
| Q00526 | CDK3 | Cyclin-dependent kinase 3 | Homo sapiens (Human) | PR |
| Q96Q40 | CDK15 | Cyclin-dependent kinase 15 | Homo sapiens (Human) | PR |
| O94921 | CDK14 | Cyclin-dependent kinase 14 | Homo sapiens (Human) | PR |
| Q00537 | CDK17 | Cyclin-dependent kinase 17 | Homo sapiens (Human) | PR |
| P49336 | CDK8 | Cyclin-dependent kinase 8 | Homo sapiens (Human) | PR |
| P50750 | CDK9 | Cyclin-dependent kinase 9 | Homo sapiens (Human) | PR |
| Q5MAI5 | CDKL4 | Cyclin-dependent kinase-like 4 | Homo sapiens (Human) | PR |
| Q00532 | CDKL1 | Cyclin-dependent kinase-like 1 | Homo sapiens (Human) | PR |
| Q92772 | CDKL2 | Cyclin-dependent kinase-like 2 | Homo sapiens (Human) | PR |
| Q8IZL9 | CDK20 | Cyclin-dependent kinase 20 | Homo sapiens (Human) | PR |
| P21127 | CDK11B | Cyclin-dependent kinase 11B | Homo sapiens (Human) | PR |
| Q8R3L8 | Cdk8 | Cyclin-dependent kinase 8 | Mus musculus (Mouse) | PR |
| Q8BWD8 | Cdk19 | Cyclin-dependent kinase 19 | Mus musculus (Mouse) | PR |
| Q6P3N6 | cdk8 | Cyclin-dependent kinase 8 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q8JH47 | cdk8 | Cyclin-dependent kinase 8 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDYDFKAKLA | AERERVEDLF | EYEGCKVGRG | TYGHVYKARR | KDGKDEKEYA | LKQIEGTGIS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| MSACREIALL | RELKHPNVIA | LQKVFLSHSD | RKVWLLFDYA | EHDLWHIIKF | HRASKANKKP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MQLPRSMVKS | LLYQILDGIH | YLHANWVLHR | DLKPANILVM | GEGPERGRVK | IADMGFARLF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NSPLKPLADL | DPVVVTFWYR | APELLLGARH | YTKAIDIWAI | GCIFAELLTS | EPIFHCRQED |
| 250 | 260 | 270 | 280 | 290 | 300 |
| IKTSNPFHHD | QLDRIFSVMG | FPADKDWEDI | RKMPEYPTLQ | KDFRRTTYAN | SSLIKYMEKH |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KVKPDSKVFL | LLQKLLTMDP | TKRITSEQAL | QDPYFQEDPL | PTLDVFAGCQ | IPYPKREFLN |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EDDPEEKGDK | NQQQQQNQHQ | QPTAPPQQAA | APPQAPPPQQ | NSTQTNGTAG | GAGAGVGGTG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AGLQHSQDSS | LNQVPPNKKP | RLGPSGANSG | GPVMPSDYQH | SSSRLNYQSS | VQGSSQSQST |
| 490 | 500 | ||||
| LGYSSSSQQS | SQYHPSHQAH | RY |