Q9ASQ5
Gene name |
CRCK3 (At2g11520, F14P14.15) |
Protein name |
Calmodulin-binding receptor-like cytoplasmic kinase 3 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT2G11520 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
367-374 (Activation loop from InterPro)
Target domain |
225-499 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
386-393 (Activation loop from InterPro)
Target domain |
225-499 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
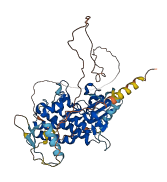
Autoinhibited structure

Activated structure

1 structures for Q9ASQ5
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q9ASQ5-F1 | Predicted | AlphaFoldDB |
63 variants for Q9ASQ5
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH05380212 | 19 | L>M | No | 1000Genomes | |
| ENSVATH12719442 | 23 | L>S | No | 1000Genomes | |
| tmp_2_4619216_A_C | 24 | Q>H | No | 1000Genomes | |
| ENSVATH12719443 | 33 | S>P | No | 1000Genomes | |
| ENSVATH00219900 | 36 | S>N | No | 1000Genomes | |
| ENSVATH05380215 | 42 | S>C | No | 1000Genomes | |
| ENSVATH00219901 | 44 | H>Y | No | 1000Genomes | |
| ENSVATH01732926 | 56 | L>W | No | 1000Genomes | |
| tmp_2_4619314_T_A | 57 | F>Y | No | 1000Genomes | |
| tmp_2_4619322_A_C | 60 | N>H | No | 1000Genomes | |
| tmp_2_4619328_A_C | 62 | N>H | No | 1000Genomes | |
| ENSVATH05380216 | 63 | P>L | No | 1000Genomes | |
| tmp_2_4619331_C_T | 63 | P>S | No | 1000Genomes | |
| ENSVATH00219902 | 66 | K>R | No | 1000Genomes | |
| ENSVATH12719444 | 80 | G>D | No | 1000Genomes | |
| ENSVATH05380220 | 90 | D>E | No | 1000Genomes | |
| ENSVATH05380221 | 95 | H>N | No | 1000Genomes | |
| ENSVATH05380222 | 95 | H>P | No | 1000Genomes | |
| ENSVATH05380226 | 101 | R>S | No | 1000Genomes | |
| ENSVATH14275310 | 104 | E>K | No | 1000Genomes | |
| ENSVATH05380227 | 108 | V>I | No | 1000Genomes | |
| ENSVATH05380229 | 109 | K>R | No | 1000Genomes | |
| tmp_2_4619567_A_C | 113 | N>T | No | 1000Genomes | |
| tmp_2_4619566_A_T | 113 | N>Y | No | 1000Genomes | |
| ENSVATH05380232 | 120 | Y>H | No | 1000Genomes | |
| ENSVATH14275311 | 123 | V>D | No | 1000Genomes | |
| tmp_2_4619602_G_C | 125 | V>L | No | 1000Genomes | |
| tmp_2_4619607_C_A | 126 | S>R | No | 1000Genomes | |
| ENSVATH12719498 | 128 | A>V | No | 1000Genomes | |
| ENSVATH00219903 | 129 | G>A | No | 1000Genomes | |
| tmp_2_4619653_C_T | 142 | P>S | No | 1000Genomes | |
| ENSVATH05380234 | 146 | K>R | No | 1000Genomes | |
| tmp_2_4619854_C_G | 180 | Q>E | No | 1000Genomes | |
| tmp_2_4619875_C_G | 187 | P>A | No | 1000Genomes | |
| tmp_2_4619876_C_T | 187 | P>L | No | 1000Genomes | |
| ENSVATH05380237 | 216 | S>G | No | 1000Genomes | |
| ENSVATH05380238 | 223 | G>D | No | 1000Genomes | |
| ENSVATH05380238 | 223 | G>V | No | 1000Genomes | |
| ENSVATH00219905 | 229 | H>N | No | 1000Genomes | |
| ENSVATH01732929 | 240 | F>Y | No | 1000Genomes | |
| ENSVATH05380239 | 243 | V>A | No | 1000Genomes | |
| ENSVATH01732930 | 243 | V>F | No | 1000Genomes | |
| ENSVATH01732930 | 243 | V>L | No | 1000Genomes | |
| ENSVATH01732931 | 246 | D>G | No | 1000Genomes | |
| ENSVATH00219906 | 247 | G>D | No | 1000Genomes | |
| ENSVATH05380240 | 247 | G>R | No | 1000Genomes | |
| ENSVATH05380240 | 247 | G>S | No | 1000Genomes | |
| ENSVATH01732935 | 293 | D>V | No | 1000Genomes | |
| tmp_2_4620609_G_A | 313 | G>S | No | 1000Genomes | |
| ENSVATH12719562 | 314 | A>T | No | 1000Genomes | |
| tmp_2_4620978_C_A | 378 | D>E | No | 1000Genomes | |
| ENSVATH01732941 | 401 | K>R | No | 1000Genomes | |
| ENSVATH01732942 | 404 | H>Q | No | 1000Genomes | |
| ENSVATH00219910 | 433 | L>P | No | 1000Genomes | |
| ENSVATH01732944 | 434 | P>H | No | 1000Genomes | |
| ENSVATH01732945 | 437 | R>K | No | 1000Genomes | |
| tmp_2_4621293_G_A | 460 | A>T | No | 1000Genomes | |
| tmp_2_4621294_C_T | 460 | A>V | No | 1000Genomes | |
| tmp_2_4621320_C_A | 469 | L>I | No | 1000Genomes | |
| ENSVATH00219913 | 478 | Q>H | No | 1000Genomes | |
| ENSVATH05380260 | 485 | K>N | No | 1000Genomes | |
| ENSVATH00219914 | 510 | E>K | No | 1000Genomes | |
| tmp_2_4621446_T_C | 511 | E>Q | No | 1000Genomes |
No associated diseases with Q9ASQ5
4 regional properties for Q9ASQ5
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 225 - 499 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 229 - 497 | IPR001245 |
| active_site | Serine/threonine-protein kinase, active site | 346 - 358 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 231 - 257 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
24 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9Y616 | IRAK3 | Interleukin-1 receptor-associated kinase 3 | Homo sapiens (Human) | PR |
| P53667 | LIMK1 | LIM domain kinase 1 | Homo sapiens (Human) | PR |
| P53671 | LIMK2 | LIM domain kinase 2 | Homo sapiens (Human) | PR |
| Q8K4B2 | Irak3 | Interleukin-1 receptor-associated kinase 3 | Mus musculus (Mouse) | PR |
| Q8CFA1 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Mus musculus (Mouse) | PR |
| O54785 | Limk2 | LIM domain kinase 2 | Mus musculus (Mouse) | PR |
| Q4QQS0 | Irak2 | Interleukin-1 receptor-associated kinase-like 2 | Rattus norvegicus (Rat) | PR |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STF0 | LECRKS3 | Receptor like protein kinase S.3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LYN6 | At3g56050 | Probable inactive receptor-like protein kinase At3g56050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VZJ9 | CRCK2 | Calmodulin-binding receptor-like cytoplasmic kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WNY5 | WAKL18 | Wall-associated receptor kinase-like 18 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94C25 | At5g20050 | Probable receptor-like protein kinase At5g20050 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LTC0 | PBL19 | Probable serine/threonine-protein kinase PBL19 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FIL7 | CRCK1 | Calmodulin-binding receptor-like cytoplasmic kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FLW0 | At5g24010 | Probable receptor-like protein kinase At5g24010 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCZ4 | FER | Receptor-like protein kinase FERONIA | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGGDDLSFTR | LVITALFGLL | MLLQIKETSA | STSFVSSSVC | KSDHLTYTKP | YQQGSLFTIN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GNPVEKLRFC | EALRFHKANG | CIFEDSFSDD | FCTIHSLLGR | RFLEEKTVKD | SKNSKPKTEY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SHVKVSIAGS | GFLLLCCALC | CPCFHKERKA | NSHEVLPKES | NSVHQVSSFE | MSPSSEKIPQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SPFRAPPSPS | RVPQSPSRYA | MSPRPSRLGP | LNLTMSQINT | ATGNFADSHQ | IGEGGFGVVF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KGVLDDGQVV | AIKRAKKEHF | ENLRTEFKSE | VDLLSKIGHR | NLVKLLGYVD | KGDERLIITE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YVRNGTLRDH | LDGARGTKLN | FNQRLEIVID | VCHGLTYLHS | YAERQIIHRD | IKSSNILLTD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SMRAKVADFG | FARGGPTDSN | QTHILTQVKG | TVGYLDPEYM | KTYHLTAKSD | VYSFGILLVE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| ILTGRRPVEA | KRLPDERITV | RWAFDKYNEG | RVFELVDPNA | RERVDEKILR | KMFSLAFQCA |
| 490 | 500 | ||||
| APTKKERPDM | EAVGKQLWAI | RSSYLRRSME |