Q99M51
Gene name |
Nck1 |
Protein name |
Cytoplasmic protein NCK1 |
Names |
NCK adaptor protein 1, Nck-1 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:17973 |
EC number |
|
Protein Class |
|
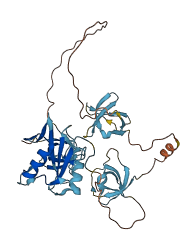
Descriptions
(Annotation based on sequence homology with O43639)
NCK2 is a cytoplasmic protein participating in the reorganization of the actin cytoskeleton and in the formation of the immunological synapses. The autoinhibitory domain locates at a conserved non-canonical (K/R)x(K/R)RxxS sequence, specifically at residues 60-110 linker between the SH3.1 and SH3.2. The autoinhibitory domain binds to the SH3.2 domain via acidic electrostatic potential on the SH3 domain surface and masks the proline-rich sequence (PRS) binding site of SH3.2 from its ligands. Only high affinity stable interactions can overcome the inhibitory interaction; such as phosphorylation of Ser85 at the interaction site.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q99M51
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q99M51-F1 | Predicted | AlphaFoldDB |
15 variants for Q99M51
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs13459245 | 4 | E>G | No | EVA | |
| rs3389064943 | 5 | V>M | No | EVA | |
| rs3389067670 | 76 | G>C | No | EVA | |
| rs3389059130 | 117 | Y>N | No | EVA | |
| rs3389042335 | 136 | M>R | No | EVA | |
| rs3389045542 | 168 | L>* | No | EVA | |
| rs3400566483 | 174 | S>C | No | EVA | |
| rs3400703994 | 176 | S>T | No | EVA | |
| rs3389053405 | 221 | E>A | No | EVA | |
| rs3389010618 | 222 | K>R | No | EVA | |
| rs3389075526 | 274 | T>S | No | EVA | |
| rs3389045554 | 328 | K>R | No | EVA | |
| rs3389032693 | 336 | E>D | No | EVA | |
| rs3389045577 | 360 | P>S | No | EVA | |
| rs3389062979 | 367 | G>R | No | EVA |
No associated diseases with Q99M51
8 regional properties for Q99M51
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SH2 domain | 280 - 376 | IPR000980 |
| domain | SH3 domain | 2 - 61 | IPR001452-1 |
| domain | SH3 domain | 106 - 165 | IPR001452-2 |
| domain | SH3 domain | 190 - 252 | IPR001452-3 |
| domain | Nck1, SH3 domain 1 | 3 - 61 | IPR035562 |
| domain | Nck1, SH3 domain 2 | 108 - 162 | IPR035564 |
| domain | Nck1, SH3 domain 3 | 193 - 249 | IPR035565 |
| domain | Nck1, SH2 domain | 280 - 376 | IPR035882 |
Functions
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein phosphatase type 1 complex | A protein complex that possesses magnesium-dependent protein serine/threonine phosphatase (AMD phosphatase) activity, and consists of a catalytic subunit and one or more regulatory subunits that dictates the phosphatase's substrate specificity, function, and activity. |
| ribosome | An intracellular organelle, about 200 A in diameter, consisting of RNA and protein. It is the site of protein biosynthesis resulting from translation of messenger RNA (mRNA). It consists of two subunits, one large and one small, each containing only protein and RNA. Both the ribosome and its subunits are characterized by their sedimentation coefficients, expressed in Svedberg units (symbol: S). Hence, the prokaryotic ribosome (70S) comprises a large (50S) subunit and a small (30S) subunit, while the eukaryotic ribosome (80S) comprises a large (60S) subunit and a small (40S) subunit. Two sites on the ribosomal large subunit are involved in translation, namely the aminoacyl site (A site) and peptidyl site (P site). Ribosomes from prokaryotes, eukaryotes, mitochondria, and chloroplasts have characteristically distinct ribosomal proteins. |
| vesicle membrane | The lipid bilayer surrounding any membrane-bounded vesicle in the cell. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ephrin receptor binding | Binding to an ephrin receptor. |
| eukaryotic initiation factor eIF2 binding | Binding to eukaryotic initiation factor eIF2, a protein complex involved in the initiation of ribosome-mediated translation. |
| molecular condensate scaffold activity | Binding and bringing together two or more macromolecules in contact, permitting those molecules to organize as a molecular condensate. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase inhibitor activity | Binds to and stops, prevents or reduces the activity of a protein kinase, an enzyme which phosphorylates a protein. |
| protein-macromolecule adaptor activity | The binding activity of a protein that brings together two or more macromolecules in contact, permitting those molecules to function in a coordinated way. The adaptor can bring together two proteins, or a protein and another macromolecule such as a lipid or a nucleic acid. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| signaling adaptor activity | The binding activity of a molecule that brings together two or more molecules in a signaling pathway, permitting those molecules to function in a coordinated way. Adaptor molecules themselves do not have catalytic activity. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
24 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| ephrin receptor signaling pathway | The series of molecular signals initiated by ephrin binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| lamellipodium assembly | Formation of a lamellipodium, a thin sheetlike extension of the surface of a migrating cell. |
| negative regulation of cell death | Any process that decreases the rate or frequency of cell death. Cell death is the specific activation or halting of processes within a cell so that its vital functions markedly cease, rather than simply deteriorating gradually over time, which culminates in cell death. |
| negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation | Any process that stops, prevents or reduces the frequency, rate or extent of endoplasmic reticulum stress-induced eiF2alpha phosphorylation. |
| negative regulation of insulin receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of insulin receptor signaling. |
| negative regulation of peptidyl-serine phosphorylation | Any process that stops, prevents, or reduces the frequency, rate or extent of the phosphorylation of peptidyl-serine. |
| negative regulation of PERK-mediated unfolded protein response | Any process that stops, prevents or reduces the frequency, rate or extent of the PERK-mediated unfolded protein response. |
| negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription from an RNA polymerase II promoter as a result of an endoplasmic reticulum stress. |
| peptidyl-serine dephosphorylation | The removal of phosphoric residues from peptidyl-O-phospho-L-serine to form peptidyl-serine. |
| positive regulation of actin filament polymerization | Any process that activates or increases the frequency, rate or extent of actin polymerization. |
| positive regulation of cap-dependent translational initiation | Any process that activates or increases the frequency, rate or extent of cap-dependent translational initiation. |
| positive regulation of cap-independent translational initiation | Any process that activates or increases the frequency, rate or extent of cap-independent translational initiation. |
| positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | Any process that activates or increases the frequency, rate or extent of an endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| positive regulation of T cell proliferation | Any process that activates or increases the rate or extent of T cell proliferation. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of translation in response to endoplasmic reticulum stress | Any process that activates, or increases the frequency, rate or extent of translation as a result of endoplasmic reticulum stress. |
| regulation of cell migration | Any process that modulates the frequency, rate or extent of cell migration. |
| response to other organism | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| substrate-dependent cell migration, cell extension | The formation of a cell surface protrusion, such as a lamellipodium or filopodium, at the leading edge of a migrating cell. |
| T cell activation | The change in morphology and behavior of a mature or immature T cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific. |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O43639 | NCK2 | Cytoplasmic protein NCK2 | Homo sapiens (Human) | EV |
| P16333 | NCK1 | Cytoplasmic protein NCK1 | Homo sapiens (Human) | PR |
| O55033 | Nck2 | Cytoplasmic protein NCK2 | Mus musculus (Mouse) | SS |
| Q64010 | Crk | Adapter molecule crk | Mus musculus (Mouse) | EV SS |
| P47941 | Crkl | Crk-like protein | Mus musculus (Mouse) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAEEVVVVAK | FDYVAQQEQE | LDIKKNERLW | LLDDSKSWWR | VRNSMNKTGF | VPSNYVERKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SARKASIVKN | LKDTLGIGKV | KRKPSVPDTA | SPADDSFVDP | GERLYDLNMP | AFVKFNYMAE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| REDELSLIKG | TKVIVMEKCS | DGWWRGSYNG | QIGWFPSNYV | TEEGDSPLGD | HVGSLSEKLA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| AVVNNLNTGQ | VLHVVQALYP | FSSSNDEELN | FEKGDVMDVI | EKPENDPEWW | KCRKINGMVG |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LVPKNYVTIM | QNNPLTSGLE | PSPPQCDYIR | PSLTGKFAGN | PWYYGKVTRH | QAEMALNERG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HEGDFLIRDS | ESSPNDFSVS | LKAQGKNKHF | KVQLKETVYC | IGQRKFSTME | ELVEHYKKAP |
| 370 | |||||
| IFTSEQGEKL | YLVKHLS |