Q99963
Gene name |
SH3GL3 (CNSA3, SH3D2C) |
Protein name |
Endophilin-A3 |
Names |
EEN-B2, Endophilin-3, SH3 domain protein 2C, SH3 domain-containing GRB2-like protein 3 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6457 |
EC number |
|
Protein Class |
|
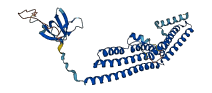
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

3 structures for Q99963
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2EW3 | NMR | - | A | 285-345 | PDB |
| 2Z0V | X-ray | 249 A | A/B | 24-256 | PDB |
| AF-Q99963-F1 | Predicted | AlphaFoldDB |
247 variants for Q99963
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA393419126 rs1473168998 |
2 | S>L | No |
ClinGen TOPMed |
|
|
CA393419133 rs1447951274 |
4 | A>T | No |
ClinGen gnomAD |
|
|
rs756581149 CA7704241 |
5 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1457828090 CA393419153 |
7 | K>R | No |
ClinGen gnomAD |
|
|
rs1331892701 CA393419197 |
13 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA393419327 rs1455824561 |
17 | F>L | No |
ClinGen TOPMed |
|
| TCGA novel | 18 | S>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7704283 rs754427453 |
22 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs1258203824 CA393419462 |
24 | A>V | No |
ClinGen gnomAD |
|
|
CA393419503 rs1252140228 |
27 | T>A | No |
ClinGen gnomAD |
|
| TCGA novel | 28 | K>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs572061818 CA274157943 |
30 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs369784464 COSM1651282 COSM965837 CA7704290 |
31 | D>H | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA7704289 rs369784464 COSM965836 COSM1651283 |
31 | D>N | Variant assessed as Somatic; 0.0001386 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 32 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs749284676 CA7704292 |
33 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA274157944 rs142447916 |
35 | D>G | No |
ClinGen ESP TOPMed |
|
|
rs768434755 CA7704293 |
36 | M>T | No |
ClinGen ExAC gnomAD |
|
| rs1412436667 | 38 | R>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 39 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA393419898 rs1566991506 |
40 | I>M | No |
ClinGen Ensembl |
|
|
rs1257749946 CA393419896 |
40 | I>T | No |
ClinGen Ensembl |
|
|
CA7704312 rs753525950 |
42 | V>F | No |
ClinGen ExAC gnomAD |
|
|
CA7704313 rs753525950 |
42 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA7704314 rs533824738 |
44 | N>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs890497772 CA274158588 |
46 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
CA393419946 rs1198536146 |
48 | A>E | No |
ClinGen gnomAD |
|
|
CA393419962 rs1459220376 |
50 | I>S | No |
ClinGen TOPMed gnomAD |
|
|
CA393419958 rs1413358891 |
50 | I>V | No |
ClinGen gnomAD |
|
|
rs1157115400 CA393419965 |
51 | L>V | No |
ClinGen gnomAD |
|
|
rs748072490 CA393419973 |
52 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA7704315 rs748072490 |
52 | S>L | No |
ClinGen ExAC gnomAD |
|
|
rs1458648566 CA393419971 |
52 | S>P | No |
ClinGen gnomAD |
|
| TCGA novel | 54 | T>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7704316 rs757962311 |
55 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757962311 CA393419992 |
55 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1386032892 CA393420006 |
57 | Y>C | No |
ClinGen gnomAD |
|
|
CA393420010 rs777243734 |
58 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs777243734 CA7704317 |
58 | L>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 61 | N>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7704345 rs761916212 |
65 | R>T | No |
ClinGen ExAC gnomAD |
|
|
rs1450117608 CA393420082 |
67 | K>E | No |
ClinGen gnomAD |
|
|
CA393420089 rs1300624896 |
68 | L>I | No |
ClinGen gnomAD |
|
|
rs1596278970 CA393420091 |
68 | L>Q | No |
ClinGen Ensembl |
|
|
rs933280142 CA274158956 |
72 | N>S | No |
ClinGen TOPMed |
|
|
CA7704346 rs767630343 COSM284961 |
75 | S>L | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA7704348 rs760929318 |
78 | R>* | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 80 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA274158959 rs1045753957 |
80 | Q>L | No |
ClinGen TOPMed |
|
|
rs1212845008 CA393420174 |
81 | V>A | No |
ClinGen gnomAD |
|
|
rs764980950 CA393420171 |
81 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7704349 rs764980950 |
81 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 83 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs763965005 CA7704352 |
85 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs373653306 CA274158961 |
86 | Y>C | No |
ClinGen ESP gnomAD |
|
|
rs1167800379 CA393420202 |
86 | Y>H | No |
ClinGen gnomAD |
|
|
rs780744126 CA7704355 |
87 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA393420210 rs780744126 |
87 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA7704354 rs756788577 |
87 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201410848 COSM1225573 COSM3401965 CA7704357 |
89 | T>M | ovary Variant assessed as Somatic; 0.0 impact. large_intestine central_nervous_system [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
CA393420262 rs1368725037 |
95 | D>E | No |
ClinGen gnomAD |
|
|
rs1268383738 CA393420260 |
95 | D>G | No |
ClinGen TOPMed |
|
|
CA393420256 rs1479595340 |
95 | D>N | No |
ClinGen TOPMed |
|
|
rs1183000378 CA393420267 |
96 | C>Y | No |
ClinGen TOPMed |
|
|
CA274158962 rs905623692 |
97 | M>V | No |
ClinGen TOPMed |
|
|
CA7704360 rs200823232 |
98 | L>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA274158964 rs559656402 |
101 | G>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes NCI-TCGA TOPMed gnomAD |
|
COSM1649178 COSM556189 rs559656402 CA393420301 |
101 | G>W | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated 1000Genomes NCI-TCGA TOPMed gnomAD |
|
CA393420313 rs1209443424 |
103 | E>K | No |
ClinGen gnomAD |
|
|
CA7704365 rs760838377 |
105 | G>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs773311339 CA7704364 |
105 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1247109031 CA393420331 |
106 | E>K | No |
ClinGen gnomAD |
|
|
rs775279705 CA7704367 |
108 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs771095393 CA7704366 |
108 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs1424390255 CA393420355 |
109 | T>S | No |
ClinGen gnomAD |
|
| TCGA novel | 110 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs762672692 CA7704368 |
110 | F>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1206986247 CA393420380 |
111 | G>V | No |
ClinGen gnomAD |
|
|
COSM1563320 rs1452155988 CA393420386 |
112 | N>S | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
CA7704392 rs142205989 |
115 | I>V | No |
ClinGen ESP ExAC TOPMed |
|
|
CA393420409 rs1358757766 |
116 | E>K | No |
ClinGen gnomAD |
|
|
CA393420421 rs1249560699 |
117 | V>A | No |
ClinGen gnomAD |
|
|
CA7704394 rs754645507 |
120 | S>C | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 121 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7704395 rs765727657 |
121 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs1471631852 CA393420465 |
124 | M>V | No |
ClinGen gnomAD |
|
|
rs1162246464 CA393420474 |
125 | A>S | No |
ClinGen gnomAD |
|
|
rs111697945 CA274159404 |
127 | V>A | No |
ClinGen Ensembl |
|
| TCGA novel | 129 | D>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA274159405 rs1044301001 |
130 | S>C | No |
ClinGen gnomAD |
|
|
rs753057998 CA7704396 |
131 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA393420540 rs1369249522 |
135 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs927249033 CA274159406 |
137 | Q>L | No |
ClinGen TOPMed |
|
|
CA393420572 rs1188202968 |
139 | F>S | No |
ClinGen TOPMed |
|
|
rs1596287726 CA393420581 |
140 | I>M | No |
ClinGen Ensembl |
|
|
CA393420584 rs1304636351 |
141 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1304636351 CA393420582 |
141 | D>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs758844147 CA7704399 |
144 | Q>E | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 148 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7704400 rs778277707 |
148 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs747469877 CA7704401 |
150 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs1221874000 CA393420657 |
151 | L>F | No |
ClinGen TOPMed |
|
|
CA393420671 rs1446144864 |
153 | E>V | No |
ClinGen TOPMed gnomAD |
|
|
CA274159407 rs764916621 |
155 | G>E | No |
ClinGen TOPMed gnomAD |
|
|
CA7704403 rs781231570 |
155 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs757735259 CA7704422 |
157 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA7704423 rs781268362 |
158 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393420725 rs1479707115 |
160 | K>E | No |
ClinGen gnomAD |
|
|
CA393420727 rs1485711780 |
160 | K>T | No |
ClinGen Ensembl |
|
|
rs745887599 CA7704425 |
163 | G>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1463995796 CA393420752 |
164 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA7704426 rs756331470 |
164 | R>H | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 164 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs780388398 CA7704427 |
165 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs955535846 CA274159829 |
165 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs575048330 CA7704428 |
167 | D>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA393420782 rs1167004728 |
169 | D>G | No |
ClinGen Ensembl |
|
|
rs772850819 CA7704430 |
169 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 171 | K>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs746973888 CA7704432 COSM267417 |
174 | R>* | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs776295437 CA7704433 |
174 | R>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1345083081 CA393420825 |
175 | V>A | No |
ClinGen TOPMed |
|
|
CA274159831 rs987379363 |
176 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
rs908627454 CA274159832 |
178 | I>T | No |
ClinGen TOPMed |
|
|
CA7704435 rs537658440 |
179 | P>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA274159833 rs1002916092 |
179 | P>L | No |
ClinGen Ensembl |
|
|
CA7704434 rs537658440 |
179 | P>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1259808836 CA393420850 |
180 | D>H | No |
ClinGen gnomAD |
|
|
CA7704437 rs149232982 COSM1588209 COSM197131 |
181 | E>K | Variant assessed as Somatic; 0.0 impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs764563470 CA7704438 |
182 | E>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA393420869 rs144447108 |
182 | E>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA393420868 rs1490020621 |
182 | E>V | No |
ClinGen gnomAD |
|
|
rs1473624747 CA393420877 |
184 | R>G | No |
ClinGen gnomAD |
|
|
rs762317932 CA7704440 |
185 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA393420884 rs762317932 |
185 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
rs767942529 CA7704441 |
186 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1178504246 CA393420896 |
187 | V>I | No |
ClinGen TOPMed |
|
|
CA393420904 rs1363459440 |
188 | E>A | No |
ClinGen gnomAD |
|
|
COSM1708468 COSM1708467 CA7704443 rs756241846 |
188 | E>K | skin [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA7704444 rs779974873 |
194 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs1055687374 CA274159834 |
196 | L>F | No |
ClinGen TOPMed |
|
|
rs755115801 CA7704446 |
200 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA7704447 rs777646810 |
200 | S>I | No |
ClinGen ExAC gnomAD |
|
|
rs777646810 CA393420991 |
200 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA7704448 rs746744566 |
201 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA393420996 rs746744566 |
201 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs770998694 CA7704449 |
204 | F>V | No |
ClinGen ExAC gnomAD |
|
|
rs1489969922 CA393421027 |
205 | L>V | No |
ClinGen gnomAD |
|
|
CA393421228 rs1596312395 |
209 | V>G | No |
ClinGen Ensembl |
|
|
rs1183626082 CA393421255 |
213 | S>T | No |
ClinGen gnomAD |
|
|
CA7704462 rs766510412 |
214 | Q>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1188971749 CA393421262 |
214 | Q>P | No |
ClinGen TOPMed |
|
|
rs1189053125 CA393421291 |
218 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs991882019 CA274161003 |
219 | I>M | No |
ClinGen TOPMed |
|
|
rs142433645 CA7704464 |
219 | I>T | No |
ClinGen ESP ExAC gnomAD |
|
|
CA393421306 rs1408190657 |
221 | A>T | No |
ClinGen gnomAD |
|
|
rs752952889 CA7704466 |
222 | A>E | No |
ClinGen ExAC gnomAD |
|
|
rs779211967 CA7704465 |
222 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA274161005 rs145979773 |
223 | L>I | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA7704468 rs781100806 |
224 | D>A | No |
ClinGen ExAC |
|
|
CA7704469 rs745846494 |
225 | Y>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7704471 rs374787134 |
226 | H>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA393421340 rs748692315 COSM3667783 COSM3667784 |
226 | H>Q | liver [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs374787134 CA7704470 |
226 | H>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs1021145699 CA274161006 |
228 | Q>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA7704473 rs768203060 |
229 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA393421356 rs1364484154 |
229 | S>T | No |
ClinGen gnomAD |
|
|
CA393421366 rs1272951900 |
230 | T>I | No |
ClinGen gnomAD |
|
| TCGA novel | 231 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs943579648 CA274161009 |
235 | E>D | No |
ClinGen TOPMed |
|
|
CA393421432 rs1261334709 |
240 | L>V | No |
ClinGen gnomAD |
|
|
CA7704475 rs761425897 |
241 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
rs1209575353 CA393421453 |
243 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1209575353 CA393421452 |
243 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
COSM3786581 rs772476093 CA7704476 COSM3786580 |
243 | R>Q | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1341513149 CA393421468 |
244 | I>L | No |
ClinGen TOPMed |
|
| TCGA novel | 247 | A>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA393421500 rs1410534845 |
249 | S>G | No |
ClinGen gnomAD |
|
|
CA393421509 rs1281341299 |
250 | V>I | No |
ClinGen gnomAD |
|
|
COSM2156481 rs765453714 COSM2156480 CA7704501 |
253 | R>* | Variant assessed as Somatic; 0.0 impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs765453714 CA7704500 |
253 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs373378258 CA7704502 |
253 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs114613246 CA7704504 |
254 | E>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA393421528 rs1268101692 |
254 | E>K | No |
ClinGen gnomAD |
|
|
rs186125782 CA7704505 |
255 | Y>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1474761474 CA393421543 |
256 | K>E | No |
ClinGen TOPMed |
|
|
CA7704506 rs766169231 |
256 | K>M | No |
ClinGen ExAC gnomAD |
|
|
rs962234369 CA274161187 |
258 | R>K | No |
ClinGen Ensembl |
|
|
CA7704510 rs778427582 |
268 | N>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA393421637 rs117673766 |
270 | V>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7704511 rs117673766 |
270 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1221721572 CA393421649 |
272 | T>A | No |
ClinGen TOPMed |
|
| TCGA novel | 273 | T>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7704514 rs746667690 COSM3377732 COSM3377731 |
278 | T>M | Variant assessed as Somatic; 0.0 impact. pancreas [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA7704516 rs781595917 |
279 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA393419238 rs1431662727 |
280 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA7704546 rs759384239 |
282 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs765011085 CA7704547 |
284 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA393419262 COSM233352 rs765011085 |
284 | P>S | skin [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA393419282 rs752670305 |
286 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA274164320 rs368195801 |
287 | Q>L | No |
ClinGen TOPMed |
|
|
CA274164319 rs368195801 |
287 | Q>R | No |
ClinGen TOPMed |
|
|
rs368496383 CA274164321 |
291 | R>C | No |
ClinGen ESP gnomAD |
|
|
CA274164322 rs764222983 |
291 | R>H | No |
ClinGen gnomAD |
|
|
rs1567042802 CA393419356 |
294 | Y>C | No |
ClinGen Ensembl |
|
|
rs139413984 CA274164323 |
295 | D>N | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA393419367 rs1451084070 |
295 | D>V | No |
ClinGen TOPMed |
|
|
CA393419383 rs1236392914 |
297 | E>K | No |
ClinGen gnomAD |
|
|
CA393419385 rs1236392914 |
297 | E>Q | No |
ClinGen gnomAD |
|
|
CA7704551 rs751036862 |
299 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA7704553 rs149974045 |
301 | Q>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7704552 rs149974045 |
301 | Q>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs750100781 CA7704554 |
301 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs756589925 CA7704555 |
303 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1489985398 CA393419453 |
303 | E>K | Variant assessed as Somatic; 4.619e-05 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA7704556 rs780546979 |
304 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs867952906 CA274164324 |
306 | F>S | No |
ClinGen Ensembl |
|
|
CA393419578 rs1481529239 |
314 | L>I | No |
ClinGen TOPMed |
|
| TCGA novel | 315 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA393419607 rs1426361758 |
316 | N>S | No |
ClinGen gnomAD |
|
|
CA393419604 rs1165177333 |
316 | N>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA7704559 rs371368722 |
317 | Q>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 317 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA274164326 rs778865546 |
317 | Q>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7704560 COSM1375022 rs778865546 |
317 | Q>R | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1284754248 CA393419663 |
321 | N>Y | No |
ClinGen TOPMed |
|
|
CA393419721 RCV000995408 rs1473247197 |
325 | G>E | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA7704561 rs748202631 |
326 | M>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1010394128 CA274164327 |
327 | I>M | No |
ClinGen TOPMed |
|
|
CA7704563 rs142360966 |
329 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 330 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs894935428 CA274164328 |
331 | S>W | No |
ClinGen Ensembl |
|
|
CA393419770 rs1247185905 COSM1265496 |
332 | G>A | oesophagus [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1215179759 CA393419773 |
333 | F>L | No |
ClinGen TOPMed |
|
|
rs1294381652 CA393419776 |
333 | F>S | No |
ClinGen gnomAD |
|
|
rs1215179759 CA393419774 |
333 | F>V | No |
ClinGen TOPMed |
|
|
rs1222374119 CA393419780 |
334 | F>I | No |
ClinGen gnomAD |
|
|
rs1222374119 CA393419781 |
334 | F>L | No |
ClinGen gnomAD |
|
|
rs1555424607 CA393419790 |
335 | P>S | No |
ClinGen Ensembl |
|
|
CA393419794 rs1481677769 |
336 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA7704568 rs762906180 |
338 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7704569 rs376114446 COSM965849 COSM1588205 |
339 | V>M | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs750152146 CA7704572 |
342 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs766883664 CA274164330 CA393419841 |
343 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7704574 COSM117872 rs766883664 |
343 | V>M | ovary Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs754191417 CA7704575 |
346 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1356162634 CA393419870 |
347 | Q>H | No |
ClinGen gnomAD |
No associated diseases with Q99963
Functions
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| acrosomal vesicle | A structure in the head of a spermatozoon that contains acid hydrolases, and is concerned with the breakdown of the outer membrane of the ovum during fertilization. It lies just beneath the plasma membrane and is derived from the lysosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| early endosome membrane | The lipid bilayer surrounding an early endosome. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| postsynaptic density, intracellular component | A network of proteins adjacent to the postsynaptic membrane forming an electron dense disc. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize neurotransmitter receptors in the adjacent membrane, such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| postsynaptic endosome | An endosomal compartment that is part of the post-synapse. Only early and recycling endosomes are typically present in the postsynapse. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| identical protein binding | Binding to an identical protein or proteins. |
| lipid binding | Binding to a lipid. |
| protein C-terminus binding | Binding to a protein C-terminus, the end of a peptide chain at which the 1-carboxyl function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| central nervous system development | The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain and spinal cord. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord. |
| negative regulation of clathrin-dependent endocytosis | Any process that stops, prevents or reduces the frequency, rate or extent of clathrin-mediated endocytosis. |
| positive regulation of neuron differentiation | Any process that activates or increases the frequency, rate or extent of neuron differentiation. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| synaptic vesicle uncoating | The removal of the protein coat on a synaptic vesicle following the pinching step at the end of budding from the presynaptic membrane. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJA1 | SH3GL1 | Endophilin-A2 | Bos taurus (Bovine) | PR |
| Q8AXV1 | SH3GL2 | Endophilin-A1 | Gallus gallus (Chicken) | PR |
| Q8AXU9 | SH3GL3 | Endophilin-A3 | Gallus gallus (Chicken) | PR |
| Q5HYK7 | SH3D19 | SH3 domain-containing protein 19 | Homo sapiens (Human) | PR |
| Q9Y371 | SH3GLB1 | Endophilin-B1 | Homo sapiens (Human) | PR |
| Q99962 | SH3GL2 | Endophilin-A1 | Homo sapiens (Human) | PR |
| Q99961 | SH3GL1 | Endophilin-A2 | Homo sapiens (Human) | PR |
| Q9Y5K6 | CD2AP | CD2-associated protein | Homo sapiens (Human) | SS |
| Q96B97 | SH3KBP1 | SH3 domain-containing kinase-binding protein 1 | Homo sapiens (Human) | EV |
| Q62419 | Sh3gl1 | Endophilin-A2 | Mus musculus (Mouse) | PR |
| Q62420 | Sh3gl2 | Endophilin-A1 | Mus musculus (Mouse) | PR |
| Q62421 | Sh3gl3 | Endophilin-A3 | Mus musculus (Mouse) | PR |
| Q9JK48 | Sh3glb1 | Endophilin-B1 | Mus musculus (Mouse) | PR |
| O35179 | Sh3gl2 | Endophilin-A1 | Rattus norvegicus (Rat) | PR |
| O35180 | Sh3gl3 | Endophilin-A3 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSVAGLKKQF | HKASQLFSEK | ISGAEGTKLD | DEFLDMERKI | DVTNKVVAEI | LSKTTEYLQP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NPAYRAKLGM | LNTVSKIRGQ | VKTTGYPQTE | GLLGDCMLKY | GKELGEDSTF | GNALIEVGES |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MKLMAEVKDS | LDINVKQTFI | DPLQLLQDKD | LKEIGHHLKK | LEGRRLDYDY | KKKRVGKIPD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EEVRQAVEKF | EESKELAERS | MFNFLENDVE | QVSQLAVFIE | AALDYHRQST | EILQELQSKL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QMRISAASSV | PRREYKPRPV | KRSSSELNGV | STTSVVKTTG | SNIPMDQPCC | RGLYDFEPEN |
| 310 | 320 | 330 | 340 | ||
| QGELGFKEGD | IITLTNQIDE | NWYEGMIHGE | SGFFPINYVE | VIVPLPQ |