Q99962
Gene name |
SH3GL2 (CNSA2, SH3D2A) |
Protein name |
Endophilin-A1 |
Names |
EEN-B1, Endophilin-1, SH3 domain protein 2A, SH3 domain-containing GRB2-like protein 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6456 |
EC number |
|
Protein Class |
|
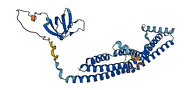
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for Q99962
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1X03 | X-ray | 310 A | A | 1-247 | PDB |
| 1X04 | X-ray | 290 A | PDB | ||
| 2D4C | X-ray | 240 A | A/B/C/D | 1-247 | PDB |
| 2DBM | NMR | - | A | 293-352 | PDB |
| AF-Q99962-F1 | Predicted | AlphaFoldDB |
273 variants for Q99962
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| TCGA novel | 4 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1244481098 CA373100357 |
4 | A>V | No |
ClinGen TOPMed |
|
|
CA373100365 rs1339128585 |
6 | L>F | No |
ClinGen TOPMed |
|
|
rs575767112 CA373100401 |
10 | F>L | No |
ClinGen 1000Genomes gnomAD |
|
|
CA373100413 rs1376637080 |
12 | K>* | No |
ClinGen gnomAD |
|
|
CA4998062 rs748128909 |
15 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs768839196 CA4998091 |
16 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373002452 rs949901949 |
20 | K>N | No |
ClinGen Ensembl |
|
|
CA373002462 rs1434121794 |
21 | V>F | No |
ClinGen gnomAD |
|
|
CA373002623 rs1336219759 |
31 | D>H | No |
ClinGen gnomAD |
|
|
rs1003368445 CA190369994 |
32 | D>A | No |
ClinGen TOPMed gnomAD |
|
|
rs768188380 CA4998094 |
33 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA373002723 rs1313511322 |
34 | K>I | No |
ClinGen TOPMed gnomAD |
|
|
CA4998096 rs760895777 |
36 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1188031647 CA373002803 |
37 | E>G | No |
ClinGen TOPMed |
|
|
CA372998854 rs1240758986 |
39 | K>R | No |
ClinGen gnomAD |
|
|
rs761266594 CA4998115 |
40 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA372998875 rs1284302235 |
42 | V>A | No |
ClinGen gnomAD |
|
|
rs766524994 CA4998116 |
42 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA4998120 rs765533067 |
50 | I>L | No |
ClinGen ExAC gnomAD |
|
|
rs765533067 CA4998119 |
50 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1184072165 CA372998937 |
51 | M>I | No |
ClinGen gnomAD |
|
|
rs1233705793 CA372998947 |
53 | K>E | No |
ClinGen gnomAD |
|
|
rs1260071120 CA372998955 COSM1265491 |
54 | T>A | oesophagus [Cosmic] | No |
ClinGen cosmic curated TOPMed |
| TCGA novel | 56 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs751434805 CA4998123 |
60 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs780520974 CA4998125 |
61 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA373001206 rs1563853926 |
63 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA373001220 rs1394821512 |
66 | A>T | No |
ClinGen gnomAD |
|
|
rs774104011 CA4998158 COSM182095 |
67 | K>N | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA4998159 rs574179015 |
69 | S>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs767344652 CA4998160 |
71 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs750307847 CA4998161 |
72 | N>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4998162 rs750307847 |
72 | N>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373001282 rs1299402448 |
74 | M>I | No |
ClinGen gnomAD |
|
|
CA4998163 rs765863127 |
75 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA373001291 rs1216920901 |
76 | K>E | No |
ClinGen gnomAD |
|
| TCGA novel | 77 | I>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1322577523 CA373001307 |
78 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA4998165 rs368973881 |
78 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs946475290 CA190374526 |
89 | A>G | No |
ClinGen TOPMed |
|
|
rs545737125 CA4998169 |
91 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs1461631960 CA373001413 |
94 | A>V | No |
ClinGen TOPMed |
|
|
CA190374527 rs1043446187 |
95 | E>D | No |
ClinGen TOPMed |
|
|
CA4998172 rs780087418 |
97 | M>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 101 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4998173 rs749411830 |
101 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373001479 rs1285129310 |
104 | L>F | No |
ClinGen TOPMed |
|
| TCGA novel | 107 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768862710 CA4998174 |
109 | N>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373001519 TCGA novel rs1421975607 |
109 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
NCI-TCGA ClinGen gnomAD |
|
rs1365168224 CA373001516 |
109 | N>S | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 110 | F>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1010309536 CA190374681 |
112 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
| TCGA novel | 116 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA190374682 rs755033673 |
117 | V>I | No |
ClinGen Ensembl |
|
|
CA190374683 rs746536752 |
118 | G>R | No |
ClinGen Ensembl |
|
| TCGA novel | 120 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA190374684 rs530596324 |
121 | M>V | No |
ClinGen Ensembl |
|
|
rs1438296171 CA373001610 |
122 | R>Q | No |
ClinGen gnomAD |
|
|
CA4998196 rs772230406 |
122 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1369428239 CA373001627 |
125 | S>A | No |
ClinGen TOPMed |
|
|
CA373001631 rs1219183971 |
125 | S>L | No |
ClinGen gnomAD |
|
|
rs1369428239 CA373001626 |
125 | S>P | No |
ClinGen TOPMed |
|
| TCGA novel | 127 | V>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs759546571 CA4998201 |
128 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA4998202 rs764803985 |
130 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA373001670 rs1330999376 |
131 | L>S | No |
ClinGen gnomAD |
|
|
CA373001686 rs1398371813 |
133 | I>M | No |
ClinGen gnomAD |
|
|
CA190374687 rs367593553 |
133 | I>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP NCI-TCGA |
|
rs1275197147 CA373001722 |
138 | N>I | No |
ClinGen gnomAD |
|
|
CA373001720 rs1275197147 COSM1107542 |
138 | N>T | endometrium [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA190374688 rs1019297708 |
139 | F>C | No |
ClinGen TOPMed gnomAD |
|
|
CA190374689 rs987335646 |
140 | I>T | No |
ClinGen Ensembl |
|
|
CA373001733 rs1217368507 |
140 | I>V | No |
ClinGen gnomAD |
|
|
rs762646779 CA373001750 |
142 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs762646779 CA4998205 |
142 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs371767677 CA190374690 |
143 | L>F | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs371767677 CA373001752 |
143 | L>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA373001762 rs1377202401 |
144 | Q>H | No |
ClinGen TOPMed |
|
|
CA4998206 rs763696032 |
147 | H>D | No |
ClinGen ExAC gnomAD |
|
|
rs540476775 CA4998207 |
147 | H>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA373001791 rs1435472405 |
148 | D>E | No |
ClinGen TOPMed |
|
|
rs201518421 CA4998208 |
149 | K>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs376054581 CA4998210 |
150 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs766744293 CA4998209 |
150 | D>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 150 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4998211 rs755497732 |
151 | L>F | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 151 | L>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1170019860 CA373001812 |
152 | R>K | No |
ClinGen gnomAD |
|
|
rs758639212 CA4998214 |
154 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1324003525 CA373001832 |
155 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
CA4998239 rs749264048 |
156 | H>Q | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 161 | L>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1471921718 CA373001909 |
164 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
COSM3433044 rs1180303494 CA373001915 |
165 | R>C | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA4998243 rs771751756 |
167 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1404256639 CA373001946 |
169 | D>E | No |
ClinGen gnomAD |
|
|
CA190374945 rs17853095 |
173 | K>E | No |
ClinGen Ensembl |
|
|
rs759948475 CA4998246 |
174 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs759948475 CA4998247 |
174 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373001978 rs1338357898 |
174 | R>Q | No |
ClinGen gnomAD |
|
|
rs1436198291 COSM214284 CA373002015 |
179 | P>L | Variant assessed as Somatic; impact. endometrium breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
CA373002020 rs1196948505 |
180 | D>V | No |
ClinGen TOPMed |
|
|
rs1378037734 CA373002028 |
181 | E>G | No |
ClinGen gnomAD |
|
|
CA4998251 rs751741211 |
183 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757525551 CA4998252 |
184 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1643797 rs781631698 CA4998253 |
184 | R>H | Variant assessed as Somatic; 0.0 impact. stomach [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 187 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1363432049 CA373002064 |
187 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA4998254 rs750231629 |
188 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA4998256 rs376134896 |
191 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA373002094 rs1200691846 |
191 | D>V | No |
ClinGen gnomAD |
|
| TCGA novel | 192 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373002098 rs1216304266 |
192 | E>K | No |
ClinGen gnomAD |
|
|
CA373002120 rs1176508437 |
195 | E>* | No |
ClinGen gnomAD |
|
|
rs1478286302 CA373002131 |
196 | I>N | No |
ClinGen gnomAD |
|
|
CA373002135 rs1171518111 |
197 | A>T | No |
ClinGen gnomAD |
|
|
CA373002154 rs1466875562 |
199 | S>L | No |
ClinGen gnomAD |
|
|
CA4998259 rs778252771 |
200 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs371069680 CA4998260 |
203 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4998261 rs138259584 |
204 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1327009104 CA373002196 |
205 | L>F | No |
ClinGen Ensembl |
|
| TCGA novel | 205 | L>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373002204 rs1305282768 |
206 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1479176820 CA373002199 |
206 | E>Q | No |
ClinGen Ensembl |
|
|
CA4998262 rs375439748 |
207 | M>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA190374947 rs971355071 |
208 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
| TCGA novel | 209 | I>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373002428 rs1256478484 |
209 | I>T | No |
ClinGen gnomAD |
|
|
rs374783522 CA4998303 |
209 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA373002478 rs1422826844 |
211 | Q>* | No |
ClinGen gnomAD |
|
|
CA4998304 rs748618916 |
215 | L>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373002578 rs1383696533 |
215 | L>P | No |
ClinGen gnomAD |
|
|
CA190375242 rs1051964069 |
216 | S>Y | No |
ClinGen Ensembl |
|
|
CA373002617 rs1159845276 |
217 | A>V | No |
ClinGen gnomAD |
|
|
CA373002684 rs1345111587 |
220 | Q>L | No |
ClinGen gnomAD |
|
|
rs1355359772 CA373002702 |
221 | A>G | No |
ClinGen gnomAD |
|
|
rs760865937 CA4998307 COSM382576 |
221 | A>T | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA4998308 rs149622931 |
222 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA373002738 rs1380198537 |
223 | L>V | No |
ClinGen gnomAD |
|
|
rs1395631064 CA373002751 |
224 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1427434423 CA373002785 |
225 | Y>C | No |
ClinGen TOPMed |
|
| TCGA novel | 226 | H>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1312655103 CA373002850 |
227 | K>E | No |
ClinGen gnomAD |
|
|
rs776842668 CA4998309 |
227 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs759704612 CA4998310 |
228 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA4998311 rs764960735 |
228 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373002906 rs1188675492 |
229 | A>V | No |
ClinGen TOPMed |
|
|
rs752535226 CA4998312 |
230 | V>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA373002924 rs1211642528 |
230 | V>G | No |
ClinGen gnomAD |
|
|
CA373002999 rs1197901325 |
233 | L>R | No |
ClinGen TOPMed |
|
|
rs751024344 CA4998315 |
234 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373003013 rs1239602961 |
234 | Q>P | No |
ClinGen gnomAD |
|
|
CA373003047 rs1179617714 |
236 | V>F | No |
ClinGen gnomAD |
|
|
CA4998316 rs184274084 |
237 | T>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1275388881 CA373003069 |
238 | V>I | No |
ClinGen TOPMed |
|
|
rs750052535 CA4998318 |
241 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA4998346 rs142672805 |
244 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA373003465 rs1478277748 |
244 | I>V | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 244 | I>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768458601 CA373003478 |
246 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA4998348 rs768458601 |
246 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA373003484 rs1374394931 |
247 | A>P | No |
ClinGen gnomAD |
|
|
CA373003486 rs1374394931 |
247 | A>S | No |
ClinGen gnomAD |
|
|
CA373003485 rs1374394931 |
247 | A>T | No |
ClinGen gnomAD |
|
|
rs1165189105 CA373003490 |
247 | A>V | No |
ClinGen gnomAD |
|
|
rs761788869 CA4998350 |
250 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs761788869 CA373003515 |
250 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767397662 CA4998351 |
250 | Q>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767397662 CA190375537 |
250 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1340537811 CA373003541 |
252 | R>K | No |
ClinGen gnomAD |
|
| TCGA novel | 253 | R>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs952883769 CA190375538 |
254 | E>Q | No |
ClinGen Ensembl |
|
|
CA190375539 rs907701905 |
255 | Y>* | No |
ClinGen gnomAD |
|
|
rs1230406384 CA373003599 |
256 | Q>L | No |
ClinGen gnomAD |
|
|
CA4998356 rs370117499 |
257 | P>R | No |
ClinGen ESP ExAC gnomAD |
|
|
rs148420843 CA4998355 |
257 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA373003628 rs1288764821 |
259 | P>A | No |
ClinGen gnomAD |
|
|
rs1045625441 CA190375540 |
259 | P>R | No |
ClinGen Ensembl |
|
|
rs751906805 CA4998358 |
260 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
COSM1461605 rs757775684 CA4998359 |
260 | R>Q | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed |
|
rs1473870587 CA373003653 |
261 | M>I | No |
ClinGen gnomAD |
|
|
rs1240630288 CA373003649 |
261 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
rs201913525 CA4998360 |
261 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373003662 rs1182267155 |
262 | S>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1182267155 CA373003664 |
262 | S>T | No |
ClinGen gnomAD |
|
|
rs1259789323 CA373003692 |
264 | E>D | No |
ClinGen TOPMed |
|
|
rs373328540 CA4998362 |
264 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs765215611 CA4998363 |
266 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA4998364 rs141085502 |
267 | T>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA373003720 rs1360566873 |
267 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1360566873 CA373003719 |
267 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
CA373003731 rs1316184108 |
269 | D>N | No |
ClinGen gnomAD |
|
|
CA190375541 rs774166488 |
270 | S>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774166488 CA373003746 |
270 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4998366 rs774166488 |
270 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1371291515 CA373003762 |
271 | T>I | No |
ClinGen TOPMed |
|
|
CA373003770 rs1244229766 |
272 | Q>* | No |
ClinGen gnomAD |
|
|
CA373003767 rs1244229766 |
272 | Q>E | No |
ClinGen gnomAD |
|
|
rs1312147607 CA373003790 |
273 | P>R | No |
ClinGen gnomAD |
|
|
CA4998367 rs142575982 |
274 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs146895243 CA190375542 |
275 | G>R | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA190375543 rs146895243 |
275 | G>W | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA4998368 rs150543523 |
276 | G>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs773215753 CA4998369 |
277 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs200824412 CA4998370 |
278 | S>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs200824412 CA373003838 |
278 | S>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4998371 rs139561767 |
279 | H>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA373003846 rs1242748977 |
279 | H>Y | No |
ClinGen gnomAD |
|
|
rs1176144215 CA373003862 |
280 | T>I | No |
ClinGen TOPMed |
|
|
rs776326202 CA373003871 |
281 | G>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA4998372 rs776326202 |
281 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs1469526352 CA373003894 |
283 | P>A | No |
ClinGen gnomAD |
|
|
CA373003905 rs1156307072 |
284 | K>E | No |
ClinGen gnomAD |
|
| TCGA novel | 284 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1414527312 CA373003927 |
287 | G>C | No |
ClinGen gnomAD |
|
|
rs373921180 CA4998394 |
288 | V>I | No |
ClinGen ESP ExAC gnomAD |
|
|
CA373004096 rs1490476561 |
289 | Q>R | No |
ClinGen gnomAD |
|
| TCGA novel | 290 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4998395 rs370568983 |
290 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767825414 CA4998396 |
292 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
COSM3942987 CA4998399 rs377538406 |
292 | Q>H | oesophagus [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs750835538 CA4998397 |
292 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
rs1372109079 CA373004153 |
295 | C>S | No |
ClinGen gnomAD |
|
|
rs1020453667 CA190375830 |
296 | R>* | No |
ClinGen TOPMed gnomAD |
|
|
rs370370071 CA4998401 |
296 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs370370071 CA4998400 |
296 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs752812516 CA4998403 |
297 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs779080747 CA4998402 |
297 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA4998406 rs139383722 |
300 | D>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4998405 COSM107345 rs139383722 |
300 | D>N | skin [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| TCGA novel | 302 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 303 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1198361878 CA373004207 |
304 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA373004224 rs1296645419 |
306 | E>* | No |
ClinGen TOPMed gnomAD |
|
|
rs1326062346 CA373004233 |
307 | G>E | No |
ClinGen gnomAD |
|
| TCGA novel | 307 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373004279 rs1182116635 |
313 | E>D | No |
ClinGen TOPMed |
|
|
COSM3699630 rs745825443 CA4998409 |
314 | G>D | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs375212321 CA4998408 COSM326461 |
314 | G>S | lung [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA4998412 COSM1107544 rs762492029 |
315 | D>N | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1489006766 CA373004293 |
316 | I>V | No |
ClinGen gnomAD |
|
|
CA373004301 rs1475487022 |
317 | I>V | No |
ClinGen gnomAD |
|
|
rs1186585152 CA373004311 |
318 | T>I | No |
ClinGen gnomAD |
|
|
CA190375832 rs941321083 |
320 | T>S | No |
ClinGen TOPMed |
|
|
CA373004326 rs1349696874 |
321 | N>S | No |
ClinGen TOPMed |
|
|
CA373004335 rs1444671962 |
322 | Q>H | No |
ClinGen TOPMed |
|
|
CA190375833 rs867743524 |
322 | Q>K | No |
ClinGen Ensembl |
|
|
CA190375834 rs973963316 |
322 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA4998416 COSM393851 rs766732200 |
324 | D>H | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs766732200 COSM455726 CA4998417 |
324 | D>N | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
| TCGA novel | 325 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs760110411 CA4998418 |
325 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs756544016 CA190375836 |
326 | N>K | No |
ClinGen Ensembl |
|
| TCGA novel | 326 | N>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1421661714 CA373004388 |
329 | E>D | No |
ClinGen gnomAD |
|
|
CA373004392 rs915787597 COSM1700960 |
330 | G>E | skin [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA190375837 rs915787597 |
330 | G>V | No |
ClinGen gnomAD |
|
|
CA190375838 rs781457075 |
331 | M>V | No |
ClinGen Ensembl |
|
|
rs765209610 CA4998419 |
333 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs918471387 CA190375839 |
333 | H>Y | No |
ClinGen TOPMed |
|
|
CA190375840 rs1036181347 |
334 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
CA373004420 rs1383318419 |
335 | H>Y | No |
ClinGen gnomAD |
|
|
CA373004435 rs758685786 |
337 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4998421 rs758685786 |
337 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4998422 rs778196221 |
338 | F>V | No |
ClinGen ExAC gnomAD |
|
|
rs1415016344 CA373004460 |
341 | I>N | No |
ClinGen TOPMed |
|
|
CA4998424 rs757049415 |
341 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA190375842 rs749097999 |
342 | N>S | No |
ClinGen TOPMed |
|
|
rs1243345507 CA373004513 |
349 | A>S | No |
ClinGen gnomAD |
|
|
CA373004516 rs1462099808 |
349 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1421330202 CA373004525 |
351 | P>H | No |
ClinGen gnomAD |
No associated diseases with Q99962
11 GO annotations of cellular component
| Name | Definition |
|---|---|
| anchoring junction | A cell junction that mechanically attaches a cell (and its cytoskeleton) to neighboring cells or to the extracellular matrix. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| clathrin-coated endocytic vesicle membrane | The lipid bilayer surrounding a clathrin-coated endocytic vesicle. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early endosome | A membrane-bounded organelle that receives incoming material from primary endocytic vesicles that have been generated by clathrin-dependent and clathrin-independent endocytosis; vesicles fuse with the early endosome to deliver cargo for sorting into recycling or degradation pathways. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| identical protein binding | Binding to an identical protein or proteins. |
| lipid binding | Binding to a lipid. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to brain-derived neurotrophic factor stimulus | A process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a brain-derived neurotrophic factor stimulus. |
| central nervous system development | The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain and spinal cord. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord. |
| dendrite extension | Long distance growth of a single dendrite involved in cellular development. |
| negative regulation of blood-brain barrier permeability | Any process that decreases blood-brain barrier permeability, the quality of the blood-brain barrier that allows for a controlled passage of substances (e.g. macromolecules, small molecules, ions) into and out of the brain. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| negative regulation of protein phosphorylation | Any process that stops, prevents or reduces the rate of addition of phosphate groups to amino acids within a protein. |
| neuron projection development | The process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| synaptic vesicle uncoating | The removal of the protein coat on a synaptic vesicle following the pinching step at the end of budding from the presynaptic membrane. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJA1 | SH3GL1 | Endophilin-A2 | Bos taurus (Bovine) | PR |
| Q8AXU9 | SH3GL3 | Endophilin-A3 | Gallus gallus (Chicken) | PR |
| Q8AXV1 | SH3GL2 | Endophilin-A1 | Gallus gallus (Chicken) | PR |
| Q5HYK7 | SH3D19 | SH3 domain-containing protein 19 | Homo sapiens (Human) | PR |
| Q9Y371 | SH3GLB1 | Endophilin-B1 | Homo sapiens (Human) | PR |
| Q99961 | SH3GL1 | Endophilin-A2 | Homo sapiens (Human) | PR |
| Q99963 | SH3GL3 | Endophilin-A3 | Homo sapiens (Human) | PR |
| Q9Y5K6 | CD2AP | CD2-associated protein | Homo sapiens (Human) | SS |
| Q96B97 | SH3KBP1 | SH3 domain-containing kinase-binding protein 1 | Homo sapiens (Human) | EV |
| Q62421 | Sh3gl3 | Endophilin-A3 | Mus musculus (Mouse) | PR |
| Q62419 | Sh3gl1 | Endophilin-A2 | Mus musculus (Mouse) | PR |
| Q62420 | Sh3gl2 | Endophilin-A1 | Mus musculus (Mouse) | PR |
| Q9JK48 | Sh3glb1 | Endophilin-B1 | Mus musculus (Mouse) | PR |
| O35180 | Sh3gl3 | Endophilin-A3 | Rattus norvegicus (Rat) | PR |
| O35179 | Sh3gl2 | Endophilin-A1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSVAGLKKQF | HKATQKVSEK | VGGAEGTKLD | DDFKEMERKV | DVTSRAVMEI | MTKTIEYLQP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NPASRAKLSM | INTMSKIRGQ | EKGPGYPQAE | ALLAEAMLKF | GRELGDDCNF | GPALGEVGEA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MRELSEVKDS | LDIEVKQNFI | DPLQNLHDKD | LREIQHHLKK | LEGRRLDFDY | KKKRQGKIPD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EELRQALEKF | DESKEIAESS | MFNLLEMDIE | QVSQLSALVQ | AQLEYHKQAV | QILQQVTVRL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EERIRQASSQ | PRREYQPKPR | MSLEFPTGDS | TQPNGGLSHT | GTPKPSGVQM | DQPCCRALYD |
| 310 | 320 | 330 | 340 | 350 | |
| FEPENEGELG | FKEGDIITLT | NQIDENWYEG | MLHGHSGFFP | INYVEILVAL | PH |