Q99741
Gene name |
CDC6 (CDC18L) |
Protein name |
Cell division control protein 6 homolog |
Names |
CDC6-related protein, Cdc18-related protein, HsCdc18, p62(cdc6), HsCDC6 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:990 |
EC number |
|
Protein Class |
|
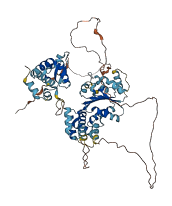
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for Q99741
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2CCH | X-ray | 170 A | E/F | 89-100 | PDB |
| 2CCI | X-ray | 270 A | F/I | 71-100 | PDB |
| 4I5L | X-ray | 243 A | B/E | 70-90 | PDB |
| 4I5N | X-ray | 280 A | B/E | 70-90 | PDB |
| AF-Q99741-F1 | Predicted | AlphaFoldDB |
423 variants for Q99741
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000374446 RCV001038572 rs776043160 |
46 | P>missing | Meier-Gorlin syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
CA8541842 rs188217621 RCV001252805 RCV002549251 |
69 | H>R | Microcephaly [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs771381953 RCV000393174 CA10645595 |
155 | T>I | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
RCV001122782 rs1156429588 CA399327891 |
170 | D>N | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
rs200468440 RCV000295424 RCV000512734 CA8541913 |
199 | Y>* | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000513028 RCV001122783 RCV000192457 rs4135010 CA205289 VAR_019349 |
238 | T>A | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA8541988 RCV000390477 rs779528702 |
282 | V>A | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001122784 RCV001521622 rs4135012 VAR_019350 RCV000116610 CA152218 |
295 | D>N | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000973558 VAR_019351 rs4135013 RCV001122786 CA152220 RCV000116611 |
299 | T>M | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs387906842 RCV000023201 CA129081 VAR_065493 |
323 | T>R | Meier-Gorlin syndrome 5 Meier-gorlin syndrome 5 (mgors5) MGORS5 [ClinVar, Ensembl, UniProt] | Yes |
ClinGen ClinVar UniProt TOPMed dbSNP |
|
rs4135016 RCV001123884 VAR_019352 CA8542058 |
378 | R>H | Meier-Gorlin syndrome 5 Variant assessed as Somatic; 0.0 impact. [ClinVar, NCI-TCGA] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
CA152214 RCV000116607 VAR_019353 rs13706 RCV000989847 RCV001519141 |
441 | V>I | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV000778495 rs372396327 CA8542166 |
519 | L>* | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA8542176 RCV001123886 rs746864516 |
531 | K>E | Meier-Gorlin syndrome 5 [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
| TCGA novel | 2 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399324502 rs1377251837 |
2 | P>T | No |
ClinGen gnomAD |
|
|
CA290494089 rs879121433 |
5 | R>P | No |
ClinGen gnomAD |
|
|
rs879121433 CA399324557 |
5 | R>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 7 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8541789 rs756261747 |
11 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs924845242 CA290494095 |
12 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA399324705 rs1467472628 |
13 | S>G | No |
ClinGen gnomAD |
|
|
rs533138014 CA399324824 |
18 | K>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs754238733 CA8541792 |
21 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs193089010 CA8541793 |
21 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs754238733 CA8541791 |
21 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 23 | L>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1291610876 CA399325031 |
31 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA399325065 rs1598512911 |
33 | K>N | No |
ClinGen Ensembl |
|
|
rs757875648 CA8541795 |
33 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA8541796 rs777176930 |
34 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399325085 rs1251654963 |
35 | E>Q | No |
ClinGen gnomAD |
|
|
CA8541797 rs746579894 |
36 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs770558092 CA8541798 |
38 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA399325146 rs1598512926 |
39 | V>A | No |
ClinGen Ensembl |
|
|
CA399325165 rs1598512933 |
41 | T>P | No |
ClinGen Ensembl |
|
|
rs1429903364 CA399325175 |
42 | V>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1415262749 CA399325172 |
42 | V>I | No |
ClinGen gnomAD |
|
|
rs1415262749 CA399325174 |
42 | V>L | No |
ClinGen gnomAD |
|
|
CA399325183 rs1171630055 |
43 | T>I | No |
ClinGen gnomAD |
|
|
CA399325189 rs1372862286 |
44 | C>S | No |
ClinGen gnomAD |
|
|
rs769593804 CA8541801 |
44 | C>W | No |
ClinGen ExAC gnomAD |
|
|
CA290494130 rs900968498 |
45 | S>Y | No |
ClinGen Ensembl |
|
|
CA399325223 rs1311115587 |
46 | P>H | No |
ClinGen gnomAD |
|
|
rs762831760 CA8541804 |
46 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8541805 rs145169276 |
47 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA399325230 rs1215372904 |
47 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs145169276 CA399325225 |
47 | R>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs866587540 CA290494157 |
48 | V>A | No |
ClinGen Ensembl |
|
|
rs1225171914 CA399325267 RCV000996531 |
50 | A>P | No |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
|
CA290494159 rs939628477 |
51 | L>P | No |
ClinGen TOPMed |
|
|
rs1282323097 CA399325304 |
52 | P>L | No |
ClinGen gnomAD |
|
|
CA8541808 rs760863872 |
54 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399325408 rs1167684753 |
58 | R>C | No |
ClinGen TOPMed |
|
|
rs375413098 CA8541809 RCV001326670 |
58 | R>H | No |
ClinGen ClinVar ESP ExAC dbSNP gnomAD |
|
|
rs765719149 CA8541811 |
59 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA8541835 rs752121916 |
60 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs1176110335 CA399325434 |
60 | G>S | No |
ClinGen gnomAD |
|
|
rs1314826623 CA399326508 |
61 | D>E | No |
ClinGen gnomAD |
|
|
rs563193240 CA290494932 |
61 | D>H | No |
ClinGen TOPMed gnomAD |
|
|
rs563193240 CA290494930 |
61 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs780716114 CA8541837 |
62 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA399326544 rs1468272767 |
63 | N>S | No |
ClinGen gnomAD |
|
|
CA399326606 rs1470782317 |
66 | N>H | No |
ClinGen gnomAD |
|
|
CA8541840 rs528888769 |
67 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA8541841 rs144462170 |
67 | T>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 69 | H>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399326679 rs1165853267 |
70 | L>V | No |
ClinGen gnomAD |
|
|
rs778655061 CA8541843 |
71 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs923461683 CA290494949 |
71 | P>S | No |
ClinGen TOPMed |
|
|
CA290494959 rs1051155107 |
72 | P>L | No |
ClinGen Ensembl |
|
|
rs1567733435 CA399326722 |
72 | P>S | No |
ClinGen Ensembl |
|
|
CA399326742 rs1286975949 |
73 | C>Y | No |
ClinGen TOPMed |
|
|
CA399326759 rs1460689160 |
74 | S>A | No |
ClinGen gnomAD |
|
|
CA290494964 rs200914287 |
79 | G>D | No |
ClinGen 1000Genomes |
|
|
COSM706478 CA399326876 rs1598513926 |
80 | K>N | lung [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA8541845 rs771094921 |
83 | N>H | No |
ClinGen ExAC gnomAD |
|
|
CA399326975 rs1278515158 |
84 | G>D | No |
ClinGen gnomAD |
|
|
CA399327009 rs1234768226 |
86 | P>A | No |
ClinGen gnomAD |
|
|
CA8541848 rs769905243 |
86 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA399327036 rs763169578 |
87 | H>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763169578 CA8541850 |
87 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1231888889 CA399327035 |
87 | H>Y | No |
ClinGen gnomAD |
|
|
rs373194490 CA8541851 RCV001348577 |
89 | H>R | No |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1267853709 CA399327128 |
91 | L>P | No |
ClinGen TOPMed |
|
|
CA399327110 rs1360683641 |
91 | L>V | No |
ClinGen TOPMed |
|
| TCGA novel | 92 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399327147 rs1486663614 |
93 | G>R | No |
ClinGen gnomAD |
|
|
CA399327157 rs1188955109 |
94 | R>* | No |
ClinGen gnomAD |
|
|
rs762281243 CA8541853 COSM3421517 |
94 | R>Q | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs201343641 CA8541854 |
100 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs551481580 CA8541855 |
102 | L>Q | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1421388749 CA399327280 |
103 | T>A | No |
ClinGen gnomAD |
|
|
CA399327301 rs1410471644 |
104 | I>M | No |
ClinGen gnomAD |
|
|
CA399327298 rs1424502353 |
104 | I>T | No |
ClinGen TOPMed |
|
|
rs1328037819 CA399327312 |
105 | K>T | No |
ClinGen gnomAD |
|
|
rs755685202 CA8541856 |
106 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765877868 CA8541857 |
107 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753538389 CA8541858 |
109 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA399327348 rs1312925151 |
111 | E>K | No |
ClinGen gnomAD |
|
|
rs1445347849 CA399327407 |
115 | V>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1445347849 CA399327405 |
115 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA399327418 rs1272291784 |
116 | H>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1340423683 CA399327424 |
116 | H>R | No |
ClinGen gnomAD |
|
|
CA399327415 rs1272291784 |
116 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA8541861 rs202098823 |
118 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA399327495 rs1489800886 |
120 | I>M | No |
ClinGen TOPMed |
|
|
RCV001341009 rs1350637996 CA399327502 |
121 | L>F | No |
ClinGen ClinVar dbSNP gnomAD |
|
| TCGA novel | 123 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399327541 rs1463133873 |
124 | V>F | No |
ClinGen TOPMed gnomAD |
|
|
CA8541864 rs199624822 |
127 | S>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs542255939 CA290495046 |
128 | Q>* | No |
ClinGen Ensembl |
|
| TCGA novel | 129 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8541866 rs769931275 |
130 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs1306886237 CA399327618 |
130 | I>V | No |
ClinGen TOPMed |
|
|
CA399327633 rs1475590899 |
132 | T>R | No |
ClinGen gnomAD |
|
|
rs1167298335 CA399327645 |
134 | S>A | No |
ClinGen gnomAD |
|
|
CA399327651 rs1270767075 |
135 | E>K | No |
ClinGen TOPMed |
|
|
rs1219998831 CA399327654 |
135 | E>V | No |
ClinGen TOPMed |
|
| TCGA novel | 136 | Q>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399327667 rs1598514103 |
137 | R>K | No |
ClinGen Ensembl |
|
|
rs185388100 CA8541867 |
137 | R>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA399327697 rs749559391 |
141 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8541869 rs768804725 |
142 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA8541870 rs774678201 |
143 | E>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 146 | C>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8541873 rs148004022 |
147 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs771381953 CA8541891 |
155 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs1598514219 CA399327809 |
156 | C>* | No |
ClinGen Ensembl |
|
|
rs1387769962 CA399327821 |
158 | Q>* | No |
ClinGen TOPMed |
|
|
rs776183829 CA8541892 |
159 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs776183829 CA399327827 |
159 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
rs759025419 CA8541893 |
160 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA290495199 rs925237047 |
162 | L>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1469606304 CA399327848 |
162 | L>V | No |
ClinGen gnomAD |
|
|
CA8541894 rs764916671 |
163 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399327864 rs1252342992 |
165 | N>T | No |
ClinGen TOPMed gnomAD |
|
|
rs752411446 CA8541895 |
166 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764013183 CA8541897 |
169 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs200103920 CA8541901 |
171 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA8541899 COSM1382950 rs757198932 |
171 | R>W | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA8541902 rs755013870 |
172 | L>P | No |
ClinGen ExAC |
|
|
CA8541903 rs779045142 |
173 | P>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 174 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1180455338 CA399327917 |
175 | R>G | No |
ClinGen TOPMed |
|
|
rs1329702045 CA399327952 |
179 | M>T | No |
ClinGen gnomAD |
|
|
CA290495241 rs1052267105 |
180 | D>H | No |
ClinGen Ensembl |
|
|
CA8541907 rs141698750 |
182 | I>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8541908 rs141698750 |
182 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs778078418 CA8541906 |
182 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA8541909 rs777070693 |
186 | L>* | No |
ClinGen ExAC gnomAD |
|
|
rs1335678307 CA399328019 |
188 | E>G | No |
ClinGen gnomAD |
|
|
CA399328037 rs1483940424 |
190 | I>L | No |
ClinGen TOPMed |
|
|
rs1237802519 CA399328048 |
190 | I>M | No |
ClinGen TOPMed |
|
|
rs1256296463 CA399328042 |
190 | I>S | No |
ClinGen TOPMed |
|
|
rs1316715520 CA399328082 |
193 | K>E | No |
ClinGen TOPMed |
|
|
CA8541911 rs759029287 |
194 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1197413245 CA399328118 |
195 | A>G | No |
ClinGen gnomAD |
|
|
rs1250634176 CA399328130 |
196 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
CA399328140 rs1482415331 |
197 | S>I | No |
ClinGen gnomAD |
|
|
CA399328143 rs1179609210 |
197 | S>R | No |
ClinGen gnomAD |
|
|
CA399328155 rs1598514320 |
198 | L>R | No |
ClinGen Ensembl |
|
|
CA8541912 rs769451211 |
199 | Y>D | No |
ClinGen ExAC gnomAD |
|
|
rs199662692 CA290495306 |
208 | K>T | No |
ClinGen Ensembl |
|
|
rs1463985809 CA399328303 |
209 | T>N | No |
ClinGen gnomAD |
|
| TCGA novel | 209 | T>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8541915 rs763890621 |
210 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1395896892 CA399328372 |
213 | S>N | No |
ClinGen gnomAD |
|
|
RCV001300527 rs550436409 CA8541916 |
214 | R>Q | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
RCV001058112 rs147186134 RCV000194819 CA209246 |
214 | R>W | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs767425948 CA8541917 |
216 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA290495358 rs867282909 |
219 | L>F | No |
ClinGen Ensembl |
|
|
rs754998763 CA8541919 |
220 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs750337305 CA8541918 |
220 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
rs752758392 CA8541941 |
222 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA8541943 rs764309883 |
223 | L>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1286834782 CA399329372 |
225 | G>R | No |
ClinGen gnomAD |
|
|
CA399329429 COSM1610183 rs1195984644 |
229 | I>V | liver [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA8541944 rs751753034 |
230 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs1567734527 CA399329449 |
230 | M>T | No |
ClinGen Ensembl |
|
|
rs751753034 CA290496289 |
230 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA8541947 rs746295385 |
234 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs781382361 CA8541946 |
234 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA399329530 rs1454494739 |
235 | S>F | No |
ClinGen gnomAD |
|
|
CA8541949 rs780775931 |
236 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1484923641 CA399329542 |
236 | L>W | No |
ClinGen TOPMed |
|
|
CA399329567 rs1305481262 |
238 | T>N | No |
ClinGen gnomAD |
|
|
CA399329574 rs1218178814 |
239 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA399329589 rs1235186305 |
240 | Q>* | No |
ClinGen gnomAD |
|
|
rs774003006 CA8541951 |
240 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA8541952 rs747783911 |
241 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399329639 rs1463785607 |
244 | P>T | No |
ClinGen gnomAD |
|
|
CA399329658 rs1317581545 |
245 | A>G | No |
ClinGen TOPMed |
|
|
rs771793749 CA8541953 |
245 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA290496316 rs1026209965 |
246 | I>V | No |
ClinGen Ensembl |
|
|
CA290496322 rs887127277 |
249 | E>D | No |
ClinGen Ensembl |
|
|
CA290496319 rs145734080 |
249 | E>K | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs145734080 CA399329702 |
249 | E>Q | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA8541955 rs144477511 |
250 | I>S | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1399104888 CA399329787 |
254 | E>D | No |
ClinGen TOPMed |
|
|
rs1382703928 CA399329816 |
257 | R>M | No |
ClinGen TOPMed |
|
|
CA399329829 rs1214110871 |
258 | P>S | No |
ClinGen gnomAD |
|
|
CA399329843 rs1158634496 |
259 | A>D | No |
ClinGen gnomAD |
|
|
CA8541956 rs766298116 |
260 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1455477932 CA399329886 |
262 | D>E | No |
ClinGen gnomAD |
|
|
RCV000501482 CA8541959 CA399329899 rs764112565 |
263 | M>I | No |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
|
CA8541958 rs370722747 |
263 | M>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs370722747 CA8541957 |
263 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1367355755 CA399329918 |
264 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA230974 RCV000116609 rs587780298 |
264 | M>V | No |
ClinGen ClinVar TOPMed dbSNP |
|
|
CA399329930 rs1162394160 |
265 | R>K | No |
ClinGen TOPMed |
|
|
RCV000193500 rs797045440 CA207033 |
270 | H>R | No |
ClinGen ClinVar Ensembl dbSNP |
|
| TCGA novel | 271 | M>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA399330016 rs1181831659 |
271 | M>V | No |
ClinGen gnomAD |
|
|
rs757400516 CA8541962 |
272 | T>P | No |
ClinGen ExAC gnomAD |
|
|
CA399330035 rs767841312 |
273 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767841312 CA8541963 |
273 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs750816199 CA8541964 |
274 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs565899649 CA8541965 |
275 | K>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs980839151 CA290496349 |
276 | G>D | No |
ClinGen TOPMed |
|
|
rs1219929306 CA399330070 |
278 | M>I | No |
ClinGen gnomAD |
|
|
rs1451607734 CA399330065 |
278 | M>V | No |
ClinGen gnomAD |
|
|
rs780653304 CA8541966 |
279 | I>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749831301 CA8541967 |
279 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs780653304 CA399330074 |
279 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8541986 rs147432601 |
280 | V>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8541985 rs766877771 |
280 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA290496461 rs1002709874 |
282 | V>L | No |
ClinGen Ensembl |
|
|
rs777349406 CA8541991 |
283 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1353466345 CA399330121 |
285 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 288 | Q>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1416767882 CA399330154 |
289 | L>P | No |
ClinGen gnomAD |
|
|
CA8541993 rs770610889 |
289 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8541995 rs199655307 |
290 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA8541994 rs780943114 |
290 | D>H | No |
ClinGen ExAC gnomAD |
|
|
CA399330164 rs1293928573 |
291 | S>G | No |
ClinGen gnomAD |
|
|
CA290496488 rs930964406 |
291 | S>N | No |
ClinGen TOPMed |
|
|
CA8541996 rs769616780 |
292 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs775505043 CA8541998 |
293 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775505043 CA8541997 |
293 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM373797 rs772220614 CA8541999 |
294 | Q>L | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA399330192 rs1278236007 |
295 | D>E | No |
ClinGen gnomAD |
|
|
CA399330189 rs958223657 |
295 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA290496509 rs958223657 |
295 | D>V | No |
ClinGen TOPMed gnomAD |
|
|
rs766753999 CA8542001 |
297 | L>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399330202 rs766753999 |
297 | L>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs537454091 CA290496516 |
298 | Y>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA290496521 rs4135013 |
299 | T>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1187792195 CA399330229 |
302 | E>K | No |
ClinGen gnomAD |
|
|
rs201579157 CA8542005 |
305 | W>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs550395628 CA290496526 |
307 | S>G | No |
ClinGen gnomAD |
|
|
rs553375751 CA8542006 |
307 | S>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs372249642 CA8542007 |
308 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs372249642 CA8542008 |
308 | N>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA290496537 rs1011555996 |
309 | S>F | No |
ClinGen TOPMed |
|
|
CA399330308 rs1412391886 |
309 | S>P | No |
ClinGen gnomAD |
|
|
rs753064373 CA399330330 |
310 | H>L | No |
ClinGen TOPMed |
|
|
rs753064373 CA290496540 |
310 | H>R | No |
ClinGen TOPMed |
|
|
CA399330324 rs1189461774 |
310 | H>Y | No |
ClinGen gnomAD |
|
| TCGA novel | 312 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs780891738 CA8542009 |
312 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs1257545317 CA399330373 |
313 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1257545317 CA399330375 |
313 | L>R | No |
ClinGen TOPMed gnomAD |
|
|
CA399330387 rs1435039121 |
314 | I>T | No |
ClinGen gnomAD |
|
|
CA8542024 RCV001338671 rs377241385 |
316 | I>V | No |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs908499387 CA290496687 |
317 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1322980850 CA399330520 |
319 | T>I | No |
ClinGen gnomAD |
|
|
CA8542025 rs763494471 |
320 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8542027 rs370007778 |
321 | D>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA290496718 rs370007778 |
321 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA399330581 rs387906842 |
323 | T>I | Meier-gorlin syndrome 5 (mgors5) [Ensembl] | No |
ClinGen TOPMed |
|
rs756766701 CA8542028 |
325 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs767102052 CA8542029 |
326 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA8542030 rs750116048 |
328 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1192914303 CA399330712 |
329 | R>S | No |
ClinGen TOPMed |
|
|
CA8542031 rs755829469 |
329 | R>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 331 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8542032 rs372655847 |
332 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs749081779 CA8542033 |
333 | R>T | No |
ClinGen ExAC gnomAD |
|
|
rs1191966404 CA399330850 |
335 | K>* | No |
ClinGen Ensembl |
|
|
CA8542035 rs754950697 |
336 | C>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399330967 rs1157491478 |
339 | Q>* | No |
ClinGen gnomAD |
|
|
CA399331054 rs1202951063 |
342 | N>T | No |
ClinGen TOPMed |
|
|
rs1056303546 CA290496780 |
344 | P>S | No |
ClinGen TOPMed |
|
|
CA399331148 rs1393259338 |
346 | Y>S | No |
ClinGen gnomAD |
|
|
rs894522069 CA290496789 |
348 | R>G | No |
ClinGen TOPMed |
|
|
CA399331182 rs1408156605 |
348 | R>T | No |
ClinGen gnomAD |
|
|
rs769959038 CA8542042 |
349 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs1286238191 CA399331201 |
349 | N>S | No |
ClinGen gnomAD |
|
|
rs1335292910 CA399331217 |
351 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
CA290496795 rs1011068507 |
351 | I>V | No |
ClinGen Ensembl |
|
|
CA8542043 rs148948716 |
354 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA399331245 rs1372873143 |
356 | Q>* | No |
ClinGen TOPMed |
|
|
rs746874201 CA8542045 |
358 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA8542046 rs774855800 |
358 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1482527038 CA399331270 |
360 | N>H | No |
ClinGen gnomAD |
|
|
rs1230435122 CA399331973 COSM978858 |
362 | V>A | Variant assessed as Somatic; 0.0003234 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA399331992 rs1330781770 |
363 | S>C | No |
ClinGen TOPMed |
|
|
rs1288579759 CA399332037 |
365 | D>A | No |
ClinGen TOPMed |
|
|
rs888632651 CA290497244 |
368 | L>M | No |
ClinGen TOPMed |
|
|
CA399332228 rs1297716244 |
372 | A>T | No |
ClinGen gnomAD |
|
|
rs900636702 CA290497247 |
374 | Q>K | No |
ClinGen Ensembl |
|
|
rs143357695 CA8542057 |
378 | R>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs4135016 CA399332380 |
378 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8542060 rs745993981 |
380 | V>F | No |
ClinGen ExAC |
|
|
CA399332462 rs1219012249 |
381 | S>C | No |
ClinGen gnomAD |
|
|
rs370762936 CA8542061 |
382 | A>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs759321100 CA290497283 |
382 | A>T | No |
ClinGen Ensembl |
|
|
CA8542062 rs780145730 |
383 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA8542063 rs749599543 |
384 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA8542064 rs769034468 |
388 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8542066 rs201176012 |
392 | D>N | No |
ClinGen 1000Genomes ExAC |
|
|
CA399332754 COSM706474 rs1415436932 |
394 | C>Y | lung Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
CA399333037 rs1598516994 |
397 | A>G | No |
ClinGen Ensembl |
|
|
CA8542087 rs780276180 |
398 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs768910005 CA8542089 |
406 | K>N | No |
ClinGen ExAC |
|
|
rs779231529 CA8542090 |
409 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8542091 rs779231529 |
409 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1387896040 CA399333442 |
412 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
CA8542093 rs773594569 |
414 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs1385903200 CA399333564 |
416 | E>A | No |
ClinGen TOPMed gnomAD |
|
|
CA8542095 rs770368321 |
416 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA8542094 COSM706473 rs760171810 |
416 | E>K | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs779176597 CA8542107 |
418 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748448453 CA8542109 |
420 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748448453 CA8542110 |
420 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399334436 rs1330902626 |
422 | E>V | No |
ClinGen gnomAD |
|
|
CA399334469 rs1401888286 |
424 | L>P | No |
ClinGen TOPMed |
|
|
CA8542112 rs778296051 |
427 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA290499760 rs1027668823 |
428 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1409984340 CA399334540 |
429 | V>I | No |
ClinGen gnomAD |
|
|
CA8542113 rs747502802 |
430 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399334554 rs747502802 |
430 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs370140805 CA399334584 |
432 | I>S | No |
ClinGen gnomAD |
|
|
rs370140805 CA290499775 |
432 | I>T | No |
ClinGen gnomAD |
|
|
CA290499780 rs868360976 |
434 | I>R | No |
ClinGen Ensembl |
|
|
CA290499777 rs868360976 |
434 | I>T | No |
ClinGen Ensembl |
|
|
CA290499796 rs749535961 |
436 | Q>H | No |
ClinGen Ensembl |
|
|
CA8542114 rs771509526 |
437 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399335266 rs13706 |
441 | V>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs13706 CA8542116 |
441 | V>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 443 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs775293499 CA8542117 |
444 | N>Y | No |
ClinGen ExAC gnomAD |
|
|
CA8542118 rs762758881 |
445 | R>K | No |
ClinGen ExAC |
|
|
CA399335373 rs1250836098 |
446 | M>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs763823622 CA8542119 |
446 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA8542120 rs774239490 |
446 | M>R | No |
ClinGen ExAC gnomAD |
|
|
CA8542121 rs761629858 |
447 | T>I | No |
ClinGen ExAC |
|
|
CA399335420 rs1324335562 |
449 | S>N | No |
ClinGen gnomAD |
|
|
CA399335482 rs1414518014 |
452 | G>E | No |
ClinGen gnomAD |
|
|
CA8542123 rs753914970 |
452 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA399335487 rs1414518014 |
452 | G>V | No |
ClinGen gnomAD |
|
|
rs527283258 CA8542124 |
453 | A>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA290499848 rs1015623708 |
454 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1207701234 CA399335531 |
455 | D>V | No |
ClinGen TOPMed |
|
|
rs1025057902 CA290499858 |
455 | D>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA399335572 rs1352371855 |
457 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
CA399335559 rs1306982620 |
457 | F>L | No |
ClinGen gnomAD |
|
|
rs1450695819 CA399335580 |
458 | P>A | No |
ClinGen gnomAD |
|
|
CA399335582 rs1450695819 |
458 | P>S | No |
ClinGen gnomAD |
|
|
rs1285144249 CA399335658 |
462 | K>M | No |
ClinGen gnomAD |
|
|
CA399335751 rs1598519106 |
468 | L>S | No |
ClinGen Ensembl |
|
|
CA290499859 rs970864346 |
470 | L>F | No |
ClinGen Ensembl |
|
|
rs752887851 CA8542126 CA290499860 |
471 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1214681511 CA399335772 |
471 | L>S | No |
ClinGen TOPMed |
|
|
rs758751645 CA8542127 |
472 | I>F | No |
ClinGen ExAC gnomAD |
|
|
CA8542128 rs777969967 |
472 | I>N | No |
ClinGen ExAC gnomAD |
|
|
rs758751645 CA290499864 |
472 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1212054058 CA399335830 |
479 | E>D | No |
ClinGen gnomAD |
|
|
CA399335851 rs1181627316 |
483 | G>E | No |
ClinGen gnomAD |
|
|
rs748883280 CA8542152 |
486 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs139013087 CA8542151 |
486 | Y>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8542154 rs778637145 |
488 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1269710544 CA399335907 |
489 | Y>* | No |
ClinGen gnomAD |
|
|
CA8542155 rs747843523 |
490 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs747843523 CA8542156 |
490 | S>R | No |
ClinGen ExAC gnomAD |
|
|
COSM978860 CA399335925 rs1208012507 |
492 | V>A | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs1404337897 CA399335922 |
492 | V>I | No |
ClinGen TOPMed |
|
|
rs1259109766 CA399335931 |
493 | C>Y | No |
ClinGen gnomAD |
|
|
CA8542157 rs773096521 |
494 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs760577823 CA8542158 |
494 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs760577823 CA399335938 |
494 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs1245787312 CA399335944 |
495 | K>R | No |
ClinGen gnomAD |
|
|
rs544050407 CA8542159 |
496 | Q>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA290500172 rs920446879 |
498 | V>A | No |
ClinGen Ensembl |
|
|
CA290500173 rs369546138 |
499 | A>V | No |
ClinGen ESP TOPMed |
|
|
rs775419688 CA8542160 |
500 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA399336001 rs1457222856 |
504 | S>T | No |
ClinGen gnomAD |
|
|
CA8542162 rs764165057 |
508 | S>P | No |
ClinGen ExAC |
|
| TCGA novel | 510 | S>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs866378978 CA290500199 |
510 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs751828306 CA8542163 |
510 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA8542164 rs369184043 |
512 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA399336091 rs369184043 |
512 | L>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1048211686 CA290500214 |
522 | K>T | No |
ClinGen TOPMed |
|
|
rs756673424 CA8542169 |
524 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1048470197 CA290500228 |
525 | K>* | No |
ClinGen gnomAD |
|
|
rs887222503 CA290500240 |
525 | K>M | No |
ClinGen Ensembl |
|
|
CA399336320 rs1048470197 |
525 | K>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 526 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs909309827 CA290500247 |
526 | E>G | No |
ClinGen TOPMed gnomAD |
|
|
rs753225105 CA8542170 |
527 | T>N | No |
ClinGen ExAC gnomAD |
|
|
rs778383432 CA8542172 |
528 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs577349066 CA8542174 |
528 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs778383432 COSM560910 CA8542173 |
528 | R>S | prostate [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA8542200 rs769667607 |
534 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs1404059943 CA399336589 |
536 | I>V | No |
ClinGen gnomAD |
|
|
rs1420689209 CA399336611 |
537 | E>A | No |
ClinGen TOPMed |
|
|
CA399336606 rs1598519699 |
537 | E>Q | No |
ClinGen Ensembl |
|
|
rs774472761 CA8542201 |
540 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA8542204 rs773577923 |
543 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772349840 CA8542203 |
543 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA8542205 rs369133472 |
544 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 551 | I>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs776903142 CA8542207 |
552 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA399336960 COSM1302767 rs1242607556 |
553 | N>D | Variant assessed as Somatic; impact. urinary_tract [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs542837328 CA290500438 |
554 | I>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs760067601 CA8542208 |
555 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765724334 CA8542209 |
556 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs752200069 CA8542210 |
556 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA399337102 rs1466590301 |
558 | G>E | No |
ClinGen gnomAD |
|
|
rs939825269 CA290500447 |
558 | G>R | No |
ClinGen TOPMed |
|
|
CA399337136 rs1189373384 |
560 | P>L | No |
ClinGen gnomAD |
1 associated diseases with Q99741
[MIM: 613805]: Meier-Gorlin syndrome 5 (MGORS5)
A syndrome characterized by bilateral microtia, aplasia/hypoplasia of the patellae, and severe intrauterine and postnatal growth retardation with short stature and poor weight gain. Additional clinical findings include anomalies of cranial sutures, microcephaly, apparently low-set and simple ears, microstomia, full lips, highly arched or cleft palate, micrognathia, genitourinary tract anomalies, and various skeletal anomalies. While almost all cases have primordial dwarfism with substantial prenatal and postnatal growth retardation, not all cases have microcephaly, and microtia and absent/hypoplastic patella are absent in some. Despite the presence of microcephaly, intellect is usually normal. {ECO:0000269|PubMed:21358632}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A syndrome characterized by bilateral microtia, aplasia/hypoplasia of the patellae, and severe intrauterine and postnatal growth retardation with short stature and poor weight gain. Additional clinical findings include anomalies of cranial sutures, microcephaly, apparently low-set and simple ears, microstomia, full lips, highly arched or cleft palate, micrognathia, genitourinary tract anomalies, and various skeletal anomalies. While almost all cases have primordial dwarfism with substantial prenatal and postnatal growth retardation, not all cases have microcephaly, and microtia and absent/hypoplastic patella are absent in some. Despite the presence of microcephaly, intellect is usually normal. {ECO:0000269|PubMed:21358632}. Note=The disease is caused by variants affecting the gene represented in this entry.
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| intercellular bridge | A direct connection between the cytoplasm of two cells that is formed following the completion of cleavage furrow ingression during cell division. They are usually present only briefly prior to completion of cytokinesis. However, in some cases, such as the bridges between germ cells during their development, they become stabilised. |
| mitotic spindle | A spindle that forms as part of mitosis. Mitotic and meiotic spindles contain distinctive complements of proteins associated with microtubules. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| spindle midzone | The area in the center of the spindle where the spindle microtubules from opposite poles overlap. |
| spindle pole | Either of the ends of a spindle, where spindle microtubules are organized; usually contains a microtubule organizing center and accessory molecules, spindle microtubules and astral microtubules. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| DNA replication origin binding | Binding to a DNA replication origin, a unique DNA sequence of a replicon at which DNA replication is initiated and proceeds bidirectionally or unidirectionally. |
| kinase binding | Binding to a kinase, any enzyme that catalyzes the transfer of a phosphate group. |
| nucleotide binding | Binding to a nucleotide, any compound consisting of a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the ribose or deoxyribose. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| cellular response to angiotensin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an angiotensin stimulus. Angiotensin is any of three physiologically active peptides (angiotensin II, III, or IV) processed from angiotensinogen. |
| cellular response to vasopressin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vasopressin stimulus. |
| DNA replication checkpoint signaling | A signal transduction process that contributes to a DNA replication checkpoint, that prevents the initiation of nuclear division until DNA replication is complete, thereby ensuring that progeny inherit a full complement of the genome. |
| DNA replication initiation | The process in which DNA-dependent DNA replication is started; this begins with the ATP dependent loading of an initiator complex onto the DNA, this is followed by DNA melting and helicase activity. In bacteria, the gene products that enable the helicase activity are loaded after the initial melting and in archaea and eukaryotes, the gene products that enable the helicase activity are inactive when they are loaded and subsequently activate. |
| mitotic cell cycle | Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent. |
| mitotic DNA replication checkpoint signaling | A signal transduction process that contributes to a mitotic DNA replication checkpoint. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of DNA replication | Any process that stops, prevents, or reduces the frequency, rate or extent of DNA replication. |
| positive regulation of chromosome segregation | Any process that activates or increases the frequency, rate or extent of chromosome segregation, the process in which genetic material, in the form of chromosomes, is organized and then physically separated and apportioned to two or more sets. |
| positive regulation of cyclin-dependent protein serine/threonine kinase activity | Any process that activates or increases the frequency, rate or extent of CDK activity. |
| positive regulation of cytokinesis | Any process that activates or increases the frequency, rate or extent of the division of the cytoplasm of a cell, and its separation into two daughter cells. |
| positive regulation of fibroblast proliferation | Any process that activates or increases the frequency, rate or extent of multiplication or reproduction of fibroblast cells. |
| regulation of cyclin-dependent protein serine/threonine kinase activity | Any process that modulates the frequency, rate or extent of cyclin-dependent protein serine/threonine kinase activity. |
| regulation of mitotic metaphase/anaphase transition | Any process that modulates the frequency, rate or extent of the cell cycle process in which a cell progresses from metaphase to anaphase during mitosis, triggered by the activation of the anaphase promoting complex by Cdc20/Sleepy homolog which results in the degradation of Securin. |
| traversing start control point of mitotic cell cycle | A cell cycle process by which a cell commits to entering S phase via a positive feedback mechanism between the regulation of transcription and G1 CDK activity. |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPQTRSQAQA | TISFPKRKLS | RALNKAKNSS | DAKLEPTNVQ | TVTCSPRVKA | LPLSPRKRLG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DDNLCNTPHL | PPCSPPKQGK | KENGPPHSHT | LKGRRLVFDN | QLTIKSPSKR | ELAKVHQNKI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LSSVRKSQEI | TTNSEQRCPL | KKESACVRLF | KQEGTCYQQA | KLVLNTAVPD | RLPAREREMD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VIRNFLREHI | CGKKAGSLYL | SGAPGTGKTA | CLSRILQDLK | KELKGFKTIM | LNCMSLRTAQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AVFPAIAQEI | CQEEVSRPAG | KDMMRKLEKH | MTAEKGPMIV | LVLDEMDQLD | SKGQDVLYTL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| FEWPWLSNSH | LVLIGIANTL | DLTDRILPRL | QAREKCKPQL | LNFPPYTRNQ | IVTILQDRLN |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QVSRDQVLDN | AAVQFCARKV | SAVSGDVRKA | LDVCRRAIEI | VESDVKSQTI | LKPLSECKSP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SEPLIPKRVG | LIHISQVISE | VDGNRMTLSQ | EGAQDSFPLQ | QKILVCSLML | LIRQLKIKEV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| TLGKLYEAYS | KVCRKQQVAA | VDQSECLSLS | GLLEARGILG | LKRNKETRLT | KVFFKIEEKE |
| 550 | |||||
| IEHALKDKAL | IGNILATGLP |