Q96L33
Gene name |
RHOV |
Protein name |
Rho-related GTP-binding protein RhoV |
Names |
CDC42-like GTPase 2, GTP-binding protein-like 2, Rho GTPase-like protein ARHV, Wnt-1 responsive Cdc42 homolog 2, WRCH-2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:171177 |
EC number |
|
Protein Class |
|
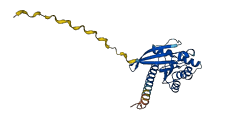
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q96L33
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q96L33-F1 | Predicted | AlphaFoldDB |
155 variants for Q96L33
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA391783744 rs1190198945 |
4 | R>Q | No |
ClinGen gnomAD |
|
|
CA391783735 rs1566863432 |
5 | E>D | No |
ClinGen Ensembl |
|
|
CA7486900 rs760568024 |
7 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA269593183 rs1037858448 |
10 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs767496320 CA7486898 |
11 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391783701 rs767496320 |
11 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391783686 rs1267670070 |
13 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs979789836 CA391783681 |
14 | L>H | No |
ClinGen TOPMed gnomAD |
|
|
rs979789836 CA269593162 |
14 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
CA7486896 rs773800967 |
16 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1368157141 CA391783664 |
17 | P>L | No |
ClinGen gnomAD |
|
|
rs916929046 CA269593146 |
19 | P>A | No |
ClinGen TOPMed |
|
|
CA7486895 rs770552675 |
20 | P>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 20 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1432552608 CA391783626 |
21 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs988533016 CA269593138 |
22 | R>Q | No |
ClinGen TOPMed |
|
|
rs1387658063 CA391783617 |
23 | R>W | No |
ClinGen gnomAD |
|
|
CA391783612 rs1458484789 |
24 | R>G | No |
ClinGen gnomAD |
|
|
CA7486894 rs748907075 |
25 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1174721561 CA391783596 |
26 | A>G | No |
ClinGen gnomAD |
|
|
CA391783600 rs1170304489 |
26 | A>T | No |
ClinGen TOPMed |
|
|
rs1174721561 CA391783595 |
26 | A>V | No |
ClinGen gnomAD |
|
|
CA391783578 rs1426437848 |
27 | P>L | No |
ClinGen gnomAD |
|
|
rs777004088 CA7486893 |
27 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391783545 rs1436895527 |
29 | E>A | No |
ClinGen TOPMed |
|
| TCGA novel | 29 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA391783386 rs1247777910 |
38 | G>V | No |
ClinGen gnomAD |
|
|
CA391783383 rs1189683859 |
39 | D>N | No |
ClinGen gnomAD |
|
|
rs1466395608 CA391783365 |
40 | G>S | No |
ClinGen gnomAD |
|
|
CA391783349 rs1254533127 |
41 | A>S | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 41 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7486891 rs747417431 |
41 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA391783337 rs1312856966 |
42 | V>M | No |
ClinGen gnomAD |
|
|
rs1226065422 CA391783314 |
43 | G>D | No |
ClinGen gnomAD |
|
|
CA391783318 rs1280582284 |
43 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA7486890 rs780683834 |
49 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs759028239 CA7486889 |
50 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA391783109 rs1214082832 |
52 | T>S | No |
ClinGen TOPMed |
|
|
rs778861445 CA7486887 |
54 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs757439908 CA7486886 |
56 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
rs1428373801 CA391782999 |
58 | A>V | No |
ClinGen gnomAD |
|
| TCGA novel | 59 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA391782945 rs1169596189 |
61 | R>Q | No |
ClinGen gnomAD |
|
|
rs755885876 CA7486883 |
64 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391782866 rs1239281442 |
66 | D>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1412778968 CA391782557 |
74 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA391782554 rs1412778968 |
74 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
rs779073916 CA269592785 |
75 | D>N | No |
ClinGen Ensembl |
|
|
CA7486854 rs775937156 |
75 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs1266646661 CA391782467 |
77 | A>S | No |
ClinGen gnomAD |
|
|
rs759988050 CA7486852 |
78 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA7486851 rs774701533 |
79 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA7486850 rs371263604 |
80 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs371263604 CA391782397 |
80 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7486849 rs749368938 |
81 | I>L | No |
ClinGen ExAC gnomAD |
|
|
rs1279054810 CA391782367 |
81 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
rs1230504623 CA391782227 |
87 | A>P | No |
ClinGen gnomAD |
|
|
rs1301226191 CA391782192 |
88 | G>E | No |
ClinGen gnomAD |
|
|
CA7486845 rs780924324 |
89 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA391782157 rs1319028797 |
89 | Q>L | No |
ClinGen gnomAD |
|
|
CA391781942 rs1596164504 |
91 | D>G | No |
ClinGen Ensembl |
|
|
rs1194133782 CA391781954 |
91 | D>H | No |
ClinGen TOPMed |
|
|
rs756808350 CA7486820 |
93 | D>V | No |
ClinGen ExAC gnomAD |
|
|
CA391781824 rs1242877755 |
96 | R>P | No |
ClinGen gnomAD |
|
|
CA7486818 rs763653151 |
100 | Y>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7486817 rs755800045 |
101 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs766790132 CA7486816 |
101 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs766790132 CA7486815 |
101 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1430105607 CA391781745 |
102 | D>N | No |
ClinGen gnomAD |
|
|
CA391781713 rs1472240692 |
104 | D>N | No |
ClinGen gnomAD |
|
|
CA269592429 rs200896433 |
107 | L>V | No |
ClinGen TOPMed |
|
|
rs1481804551 CA391781645 |
108 | A>E | No |
ClinGen gnomAD |
|
|
rs1481804551 CA391781642 |
108 | A>G | No |
ClinGen gnomAD |
|
| TCGA novel | 110 | F>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7486812 rs200872723 |
111 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs531217839 CA7486811 |
116 | S>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1369433825 CA391781508 |
116 | S>R | No |
ClinGen TOPMed |
|
|
rs1438969112 CA391781469 |
119 | Q>E | No |
ClinGen gnomAD |
|
|
rs747265102 CA7486808 |
125 | W>L | No |
ClinGen ExAC gnomAD |
|
|
CA7486807 rs775096024 |
130 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs772021008 CA7486806 |
131 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs1267767430 CA391781228 |
133 | N>I | No |
ClinGen TOPMed |
|
|
rs1335488156 CA391781225 |
133 | N>K | No |
ClinGen TOPMed |
|
|
rs774198794 CA391781209 |
134 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774198794 CA7486805 |
134 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 135 | Q>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs771062342 CA7486803 |
136 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA269592342 rs966833198 |
143 | T>I | No |
ClinGen Ensembl |
|
|
CA391781067 rs1470786549 |
144 | Q>P | No |
ClinGen gnomAD |
|
|
rs1235123996 CA391781047 |
145 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1420683111 CA391781044 |
146 | D>N | No |
ClinGen TOPMed |
|
|
CA7486798 rs755674136 |
147 | L>M | No |
ClinGen ExAC gnomAD |
|
|
rs752426836 CA7486797 |
148 | R>T | No |
ClinGen ExAC gnomAD |
|
|
CA7486796 rs780749750 |
149 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA391780979 rs1186510308 |
150 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1224451266 CA391780920 |
153 | V>A | No |
ClinGen gnomAD |
|
|
CA391780874 rs1461445419 |
156 | Q>H | No |
ClinGen TOPMed |
|
|
rs1566863115 CA391780861 |
157 | L>P | No |
ClinGen Ensembl |
|
|
rs971957892 CA269592312 |
158 | D>A | No |
ClinGen TOPMed |
|
| TCGA novel | 158 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1191427776 CA391780800 |
159 | Q>H | No |
ClinGen TOPMed |
|
|
rs963315887 CA269592311 |
159 | Q>K | No |
ClinGen TOPMed gnomAD |
|
|
CA391780804 rs1457885897 |
159 | Q>L | No |
ClinGen TOPMed |
|
|
CA7486794 rs750843485 |
160 | G>A | No |
ClinGen ExAC |
|
|
CA7486795 rs758738383 |
160 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA7486792 rs762311867 |
161 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 162 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7486791 rs753797823 |
162 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA7486790 rs764252736 |
164 | G>C | No |
ClinGen ExAC gnomAD |
|
|
CA391780737 rs1235201917 |
165 | P>S | No |
ClinGen TOPMed |
|
|
rs553339259 CA391780706 |
167 | P>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA391780698 rs1257486821 |
167 | P>L | No |
ClinGen gnomAD |
|
|
rs553339259 CA7486788 |
167 | P>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs553339259 CA391780708 |
167 | P>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1251835647 CA391780678 |
168 | Q>H | No |
ClinGen TOPMed |
|
| TCGA novel | 168 | Q>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA391780666 rs1410716300 |
169 | P>H | No |
ClinGen gnomAD |
|
|
rs1381528810 CA391780670 |
169 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1285015391 CA391780650 |
170 | Q>E | No |
ClinGen gnomAD |
|
|
rs1447695171 CA391780631 |
170 | Q>L | No |
ClinGen TOPMed gnomAD |
|
|
CA269592253 rs970895564 |
172 | Q>* | No |
ClinGen Ensembl |
|
|
CA391780554 rs1475362960 |
173 | G>V | No |
ClinGen TOPMed |
|
|
CA391780529 rs1596164328 |
175 | A>S | No |
ClinGen Ensembl |
|
|
rs1195935616 CA391780466 |
177 | K>N | No |
ClinGen gnomAD |
|
|
CA391780431 COSM961311 rs1463691429 |
179 | R>Q | Variant assessed as Somatic; 0.0 impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 182 | C>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA391780376 rs1596164322 |
182 | C>S | No |
ClinGen Ensembl |
|
|
rs1216538527 CA391780319 |
184 | L>V | No |
ClinGen gnomAD |
|
|
CA269592177 rs897574577 |
189 | L>V | No |
ClinGen Ensembl |
|
|
CA391780019 rs1566863052 |
197 | V>I | No |
ClinGen Ensembl |
|
|
CA7486781 rs777109191 |
198 | F>V | No |
ClinGen ExAC gnomAD |
|
|
rs1432661870 CA391780001 |
199 | D>V | No |
ClinGen gnomAD |
|
| TCGA novel | 200 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7486780 rs769317376 |
201 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747658164 CA7486779 |
202 | I>L | No |
ClinGen ExAC gnomAD |
|
|
rs780808040 CA7486778 |
206 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs754691420 CA7486777 |
207 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1005924481 CA269592140 |
211 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA7486776 rs750718965 |
211 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA7486774 rs757652620 |
212 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA391779870 rs1167837976 |
212 | L>Q | No |
ClinGen gnomAD |
|
|
CA269592123 rs1042751845 |
216 | L>R | No |
ClinGen TOPMed |
|
|
rs754397050 CA7486773 |
217 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs764547291 CA7486772 |
219 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1596164265 CA391779744 |
221 | V>G | No |
ClinGen Ensembl |
|
|
rs1050357314 CA269592112 |
221 | V>M | No |
ClinGen gnomAD |
|
|
rs767785165 CA7486769 |
222 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs752789848 CA7486770 |
222 | R>S | No |
ClinGen ExAC |
|
|
CA391779720 rs1596164255 |
223 | T>P | No |
ClinGen Ensembl |
|
|
rs1596164251 CA391779690 |
225 | S>P | No |
ClinGen Ensembl |
|
|
rs1471626895 CA391779669 |
226 | R>C | No |
ClinGen gnomAD |
|
|
CA391779611 rs1225818851 |
228 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA391779633 rs1282424110 |
228 | R>S | No |
ClinGen gnomAD |
|
|
CA7486767 rs774239954 |
230 | K>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 234 | C>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with Q96L33
36 regional properties for Q96L33
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | Actinin-type actin-binding domain, conserved site | 56 - 65 | IPR001589-1 |
| conserved_site | Actinin-type actin-binding domain, conserved site | 130 - 154 | IPR001589-2 |
| domain | Pleckstrin homology domain, spectrin-type | 2200 - 2219 | IPR001605-1 |
| domain | Pleckstrin homology domain, spectrin-type | 2220 - 2241 | IPR001605-2 |
| domain | Pleckstrin homology domain, spectrin-type | 2262 - 2279 | IPR001605-3 |
| domain | Pleckstrin homology domain, spectrin-type | 2282 - 2300 | IPR001605-4 |
| domain | Calponin homology domain | 54 - 158 | IPR001715-1 |
| domain | Calponin homology domain | 173 - 278 | IPR001715-2 |
| domain | Pleckstrin homology domain | 2197 - 2309 | IPR001849 |
| repeat | Spectrin repeat | 303 - 411 | IPR002017-1 |
| repeat | Spectrin repeat | 423 - 525 | IPR002017-2 |
| repeat | Spectrin repeat | 530 - 636 | IPR002017-3 |
| repeat | Spectrin repeat | 639 - 742 | IPR002017-4 |
| repeat | Spectrin repeat | 745 - 847 | IPR002017-5 |
| repeat | Spectrin repeat | 850 - 952 | IPR002017-6 |
| repeat | Spectrin repeat | 957 - 1060 | IPR002017-7 |
| repeat | Spectrin repeat | 1063 - 1166 | IPR002017-8 |
| repeat | Spectrin repeat | 1170 - 1258 | IPR002017-9 |
| repeat | Spectrin repeat | 1276 - 1376 | IPR002017-10 |
| repeat | Spectrin repeat | 1381 - 1482 | IPR002017-11 |
| repeat | Spectrin repeat | 1486 - 1590 | IPR002017-12 |
| repeat | Spectrin repeat | 1592 - 1696 | IPR002017-13 |
| repeat | Spectrin repeat | 1698 - 1801 | IPR002017-14 |
| repeat | Spectrin repeat | 1805 - 1907 | IPR002017-15 |
| repeat | Spectrin repeat | 1914 - 2014 | IPR002017-16 |
| repeat | Spectrin repeat | 2018 - 2097 | IPR002017-17 |
| repeat | Spectrin/alpha-actinin | 305 - 411 | IPR018159-1 |
| repeat | Spectrin/alpha-actinin | 425 - 525 | IPR018159-2 |
| repeat | Spectrin/alpha-actinin | 530 - 849 | IPR018159-3 |
| repeat | Spectrin/alpha-actinin | 851 - 1169 | IPR018159-4 |
| repeat | Spectrin/alpha-actinin | 1171 - 1380 | IPR018159-5 |
| repeat | Spectrin/alpha-actinin | 1381 - 1592 | IPR018159-6 |
| repeat | Spectrin/alpha-actinin | 1593 - 1805 | IPR018159-7 |
| repeat | Spectrin/alpha-actinin | 1806 - 2027 | IPR018159-8 |
| repeat | Spectrin/alpha-actinin | 2020 - 2128 | IPR018159-9 |
| domain | Pleckstrin homology domain 9 | 2201 - 2303 | IPR041681 |
Functions
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| endosome membrane | The lipid bilayer surrounding an endosome. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| metal ion binding | Binding to a metal ion. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| Cdc42 protein signal transduction | The series of molecular signals within the cell that are mediated by the Cdc42 protein switching to a GTP-bound active state. |
| endocytosis | A vesicle-mediated transport process in which cells take up external materials or membrane constituents by the invagination of a small region of the plasma membrane to form a new membrane-bounded vesicle. |
| establishment or maintenance of cell polarity | Any cellular process that results in the specification, formation or maintenance of anisotropic intracellular organization or cell growth patterns. |
36 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P61585 | RHOA | Transforming protein RhoA | Bos taurus (Bovine) | PR |
| Q1RMJ6 | RHOC | Rho-related GTP-binding protein RhoC | Bos taurus (Bovine) | PR |
| Q2HJ68 | RND1 | Rho-related GTP-binding protein Rho6 | Bos taurus (Bovine) | PR |
| Q3ZBW5 | RHOB | Rho-related GTP-binding protein RhoB | Bos taurus (Bovine) | PR |
| Q17QI8 | Rhov | Rho-related GTP-binding protein RhoV | Bos taurus (Bovine) | PR |
| Q9PSX7 | RHOC | Rho-related GTP-binding protein RhoC | Gallus gallus (Chicken) | PR |
| P48148 | Rho1 | Ras-like GTP-binding protein Rho1 | Drosophila melanogaster (Fruit fly) | PR |
| O94955 | RHOBTB3 | Rho-related BTB domain-containing protein 3 | Homo sapiens (Human) | EV |
| O94844 | RHOBTB1 | Rho-related BTB domain-containing protein 1 | Homo sapiens (Human) | PR |
| P08134 | RHOC | Rho-related GTP-binding protein RhoC | Homo sapiens (Human) | PR |
| P61586 | RHOA | Transforming protein RhoA | Homo sapiens (Human) | PR |
| P61587 | RND3 | Rho-related GTP-binding protein RhoE | Homo sapiens (Human) | PR |
| P62745 | RHOB | Rho-related GTP-binding protein RhoB | Homo sapiens (Human) | PR |
| Q92730 | RND1 | Rho-related GTP-binding protein Rho6 | Homo sapiens (Human) | PR |
| Q62159 | Rhoc | Rho-related GTP-binding protein RhoC | Mus musculus (Mouse) | PR |
| Q8BLR7 | Rnd1 | Rho-related GTP-binding protein Rho6 | Mus musculus (Mouse) | PR |
| Q9QUI0 | Rhoa | Transforming protein RhoA | Mus musculus (Mouse) | PR |
| P61588 | Rnd3 | Rho-related GTP-binding protein RhoE | Mus musculus (Mouse) | PR |
| Q9DAK3 | Rhobtb1 | Rho-related BTB domain-containing protein 1 | Mus musculus (Mouse) | PR |
| Q9CTN4 | Rhobtb3 | Rho-related BTB domain-containing protein 3 | Mus musculus (Mouse) | SS |
| P62746 | Rhob | Rho-related GTP-binding protein RhoB | Mus musculus (Mouse) | PR |
| Q8VDU1 | Rhov | Rho-related GTP-binding protein RhoV | Mus musculus (Mouse) | PR |
| O77683 | RND3 | Rho-related GTP-binding protein RhoE | Sus scrofa (Pig) | PR |
| Q6SA80 | Rnd3 | Rho-related GTP-binding protein RhoE | Rattus norvegicus (Rat) | PR |
| P62747 | Rhob | Rho-related GTP-binding protein RhoB | Rattus norvegicus (Rat) | PR |
| P61589 | Rhoa | Transforming protein RhoA | Rattus norvegicus (Rat) | PR |
| Q9Z1Y0 | Rhov | Rho-related GTP-binding protein RhoV | Rattus norvegicus (Rat) | PR |
| Q9SSX0 | RAC1 | Rac-like GTP-binding protein 1 | Oryza sativa subsp japonica (Rice) | PR |
| Q6Z808 | RAC3 | Rac-like GTP-binding protein 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q68Y52 | RAC2 | Rac-like GTP-binding protein 2 | Oryza sativa subsp japonica (Rice) | PR |
| Q67VP4 | RAC4 | Rac-like GTP-binding protein 4 | Oryza sativa subsp japonica (Rice) | PR |
| Q22038 | rho-1 | Ras-like GTP-binding protein rhoA | Caenorhabditis elegans | PR |
| Q9SU67 | ARAC8 | Rac-like GTP-binding protein ARAC8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82480 | ARAC7 | Rac-like GTP-binding protein ARAC7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38903 | ARAC2 | Rac-like GTP-binding protein ARAC2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82481 | ARAC10 | Rac-like GTP-binding protein ARAC10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPPRELSEAE | PPPLRAPTPP | PRRRSAPPEL | GIKCVLVGDG | AVGKSSLIVS | YTCNGYPARY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RPTALDTFSV | QVLVDGAPVR | IELWDTAGQE | DFDRLRSLCY | PDTDVFLACF | SVVQPSSFQN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ITEKWLPEIR | THNPQAPVLL | VGTQADLRDD | VNVLIQLDQG | GREGPVPQPQ | AQGLAEKIRA |
| 190 | 200 | 210 | 220 | 230 | |
| CCYLECSALT | QKNLKEVFDS | AILSAIEHKA | RLEKKLNAKG | VRTLSRCRWK | KFFCFV |