Q96JJ3
Gene name |
ELMO2 (CED12A, KIAA1834) |
Protein name |
Engulfment and cell motility protein 2 |
Names |
Protein ced-12 homolog A , hCed-12A |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:63916 |
EC number |
|
Protein Class |
|
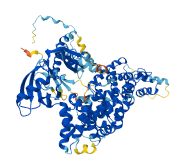
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1-113 (Ras-binding domain) |
Relief mechanism |
Partner binding |
Assay |
|
Target domain |
1-113 (Ras-binding domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Patel M et al. (2011) "The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family", The Journal of biological chemistry, 286, 38969-79
- Patel M et al. (2010) "An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling", Current biology : CB, 20, 2021-7
- Patel M et al. (2011) "Opening up on ELMO regulation: New insights into the control of Rac signaling by the DOCK180/ELMO complex", Small GTPases, 2, 268-275
Autoinhibited structure

Activated structure

3 structures for Q96JJ3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6IDX | X-ray | 170 A | A | 1-520 | PDB |
| 6IE1 | X-ray | 248 A | A | 1-520 | PDB |
| AF-Q96JJ3-F1 | Predicted | AlphaFoldDB |
549 variants for Q96JJ3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV002251070 rs2145827538 |
104 | M>missing | Primary intraosseous venous malformation [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV002244261 rs1372506599 |
483 | R>* | Primary intraosseous venous malformation [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
rs1568748859 RCV000761502 |
509 | R>* | Primary intraosseous venous malformation [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
CA10586368 rs886037919 RCV000240837 |
694 | L>missing | Primary intraosseous venous malformation [ClinVar] | Yes |
ClinGen ClinVar dbSNP |
| rs1313173820 | 2 | P>S | No | gnomAD | |
|
rs781450905 COSM3547253 |
4 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2060210543 | 4 | P>S | No | Ensembl | |
| rs1298321706 | 7 | I>V | No | Ensembl | |
| rs2145843277 | 10 | V>L | No | Ensembl | |
| rs2060210140 | 12 | I>T | No | TOPMed | |
| rs2060210204 | 12 | I>V | No | Ensembl | |
| COSM3770822 | 13 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
TCGA novel rs2060209953 |
14 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1334813316 | 14 | W>R | No |
TOPMed gnomAD |
|
| rs768673754 | 16 | G>R | No |
ExAC gnomAD |
|
| rs780710246 | 19 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA |
| rs758917154 | 21 | L>F | No |
ExAC gnomAD |
|
| rs779460586 | 25 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs370761872 | 28 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs148583859 | 28 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1170346688 | 29 | P>A | No | gnomAD | |
| rs1424847566 | 29 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1182091776 | 31 | A>V | No | gnomAD | |
| rs752239276 | 33 | I>V | No |
ExAC gnomAD |
|
| rs1427031268 | 36 | E>V | No | Ensembl | |
| rs767049111 | 39 | D>G | No |
ExAC gnomAD |
|
| rs1249170051 | 40 | G>A | No | gnomAD | |
| rs2060195175 | 42 | S>L | No | TOPMed | |
| rs1301826964 | 44 | P>S | No | gnomAD | |
| rs2060194877 | 45 | N>S | No | TOPMed | |
| rs766239453 | 46 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs758305382 | 49 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs143424054 | 50 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1600876164 | 50 | T>P | No | Ensembl | |
| rs549091525 | 51 | L>V | No |
1000Genomes ExAC gnomAD |
|
|
COSM1713580 rs775922880 |
52 | R>C | skin [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1239729184 | 52 | R>H | No | TOPMed | |
| rs202205136 | 55 | D>Y | No | 1000Genomes | |
| rs1170482925 | 56 | G>D | No | TOPMed | |
| rs1490147273 | 57 | P>L | No |
TOPMed gnomAD |
|
| rs2060193859 | 57 | P>S | No | Ensembl | |
| COSM1412183 | 60 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1348339669 | 61 | I>L | No |
TOPMed gnomAD |
|
| rs1348339669 | 61 | I>V | No |
TOPMed gnomAD |
|
| rs1260335731 | 62 | T>I | No | gnomAD | |
| rs1325737958 | 63 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1281388430 | 64 | Q>R | No | gnomAD | |
| rs1240078857 | 65 | T>I | No |
TOPMed gnomAD |
|
| rs1164888867 | 65 | T>S | No | gnomAD | |
| rs770222554 | 66 | R>C | No |
ExAC gnomAD |
|
| rs2060181190 | 66 | R>H | No | TOPMed | |
| rs2060181190 | 66 | R>L | No | TOPMed | |
| rs370462159 | 69 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2060181064 | 70 | K>E | No | Ensembl | |
| rs1368932281 | 72 | G>E | No |
TOPMed gnomAD |
|
| rs1419698700 | 74 | I>V | No |
TOPMed gnomAD |
|
| rs774088400 | 76 | Q>E | No | TOPMed | |
| COSM443936 | 76 | Q>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774088400 | 76 | Q>K | No | TOPMed | |
| rs149862820 | 79 | I>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs149862820 | 79 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs771678995 | 81 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1312652437 | 81 | P>L | No |
TOPMed gnomAD |
|
| rs771678995 | 81 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs201165179 | 83 | R>Q | No |
ExAC gnomAD |
|
| rs530112501 | 83 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs143383252 | 86 | R>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs752371094 | 86 | R>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 87 | Q>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758503208 | 89 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1401098266 | 89 | M>L | No | gnomAD | |
| rs1372825753 | 91 | R>T | No |
TOPMed gnomAD |
|
| rs2060106661 | 92 | T>N | No |
TOPMed gnomAD |
|
| rs760666181 | 94 | S>A | No | TOPMed | |
| rs1472304709 | 94 | S>L | No | gnomAD | |
| rs760666181 | 94 | S>P | No | TOPMed | |
| rs1201706901 | 96 | N>I | No | gnomAD | |
| rs894701375 | 99 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs894701375 | 99 | T>N | No |
TOPMed gnomAD |
|
| rs754408925 | 100 | R>Q | No |
ExAC gnomAD |
|
| rs187095956 | 100 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1285880351 | 103 | A>T | No | gnomAD | |
| rs761113935 | 104 | M>I | No |
ExAC gnomAD |
|
| rs2060105760 | 104 | M>T | No | TOPMed | |
| rs764441184 | 104 | M>V | No |
ExAC gnomAD |
|
| rs775666815 | 105 | K>R | No |
ExAC gnomAD |
|
| rs2060105428 | 106 | E>* | No | Ensembl | |
| rs767227832 | 107 | L>V | No |
ExAC gnomAD |
|
| rs2060105208 | 109 | K>M | No | TOPMed | |
| rs1343510124 | 109 | K>N | No | gnomAD | |
| TCGA novel | 113 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770618795 | 113 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1430245718 | 114 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs769563413 | 115 | T>I | No |
ExAC gnomAD |
|
| rs1568775177 | 116 | F>C | No | Ensembl | |
| rs142737262 | 116 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2060103865 | 117 | A>T | No | TOPMed | |
| rs754772676 | 118 | T>A | No |
ExAC gnomAD |
|
| rs1487052859 | 120 | F>L | No |
TOPMed gnomAD |
|
|
COSM3547252 rs1240780094 |
122 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1464110700 | 123 | M>I | No | gnomAD | |
| rs771697087 | 123 | M>R | No | Ensembl | |
| rs771697087 | 123 | M>T | No | Ensembl | |
| rs1418031235 | 123 | M>V | No |
TOPMed gnomAD |
|
| rs2060102798 | 124 | D>N | No | gnomAD | |
| rs779094272 | 125 | G>S | No |
ExAC gnomAD |
|
| rs757334097 | 127 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs1309972224 | 127 | I>T | No | gnomAD | |
| rs757334097 | 127 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1286830468 | 132 | L>F | No |
TOPMed gnomAD |
|
| rs200067535 | 133 | V>M | No |
1000Genomes gnomAD |
|
| rs767818738 | 134 | E>K | No |
ExAC gnomAD |
|
| TCGA novel | 137 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369135440 | 137 | T>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs369135440 | 137 | T>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1405320041 | 138 | K>E | No |
TOPMed gnomAD |
|
| rs572805895 | 139 | L>P | No |
1000Genomes ExAC gnomAD |
|
| rs762570297 | 140 | L>S | No |
ExAC gnomAD |
|
| COSM3547251 | 141 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1445669041 | 141 | S>P | No | gnomAD | |
| rs1243341035 | 142 | H>Y | No | gnomAD | |
| rs1247869989 | 144 | S>G | No |
TOPMed gnomAD |
|
| rs760484219 | 145 | E>A | No |
ExAC gnomAD |
|
| COSM724376 | 145 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs944993339 | 147 | L>Q | No | TOPMed | |
| rs2060066307 | 150 | T>P | No | Ensembl | |
| rs577555657 | 153 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2060065951 | 158 | M>I | No | Ensembl | |
| rs1307944244 | 158 | M>V | No | gnomAD | |
| rs1425779402 | 160 | H>Q | No |
TOPMed gnomAD |
|
| rs1242666889 | 160 | H>Y | No | gnomAD | |
| rs771821164 | 161 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs2060065713 | 162 | I>V | No | gnomAD | |
| rs2060065555 | 164 | S>C | No | TOPMed | |
| rs911857943 | 167 | M>I | No |
TOPMed gnomAD |
|
| rs1195967908 | 167 | M>T | No |
TOPMed gnomAD |
|
| rs1162010841 | 167 | M>V | No |
TOPMed gnomAD |
|
| rs2060065247 | 169 | S>P | No | Ensembl | |
| rs778040432 | 170 | I>V | No |
ExAC gnomAD |
|
| rs1159438280 | 171 | T>S | No | gnomAD | |
| rs748188452 | 172 | F>C | No |
ExAC gnomAD |
|
| rs950801987 | 173 | I>F | No | TOPMed | |
| COSM3547250 | 173 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2060064641 | 174 | K>E | No | Ensembl | |
| rs2060041931 | 177 | A>T | No | TOPMed | |
| rs745452765 | 178 | G>E | No | TOPMed | |
| rs745452765 | 178 | G>V | No | TOPMed | |
| rs150136023 | 183 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1843559515 | 184 | M>L | No | Ensembl | |
| rs2060041525 | 184 | M>R | No | Ensembl | |
| rs2060041299 | 185 | V>A | No | Ensembl | |
| rs2060041299 | 185 | V>E | No | Ensembl | |
| rs2060041391 | 185 | V>M | No | Ensembl | |
| rs1385925999 | 186 | D>G | No | gnomAD | |
| rs758899161 | 187 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs758899161 | 187 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs925209638 | 189 | I>M | No | TOPMed | |
| rs1475879541 | 189 | I>V | No | TOPMed | |
| rs1267939740 | 190 | L>H | No |
TOPMed gnomAD |
|
| TCGA novel | 191 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 195 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2060040455 | 195 | A>V | No | TOPMed | |
| rs1485580663 | 196 | I>F | No | gnomAD | |
| rs2060040148 | 196 | I>M | No | Ensembl | |
| rs2060040211 | 196 | I>N | No | Ensembl | |
| rs1485580663 | 196 | I>V | No | gnomAD | |
| rs372666947 | 197 | L>P | No |
ESP ExAC gnomAD |
|
| rs1373602909 | 199 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs753399070 | 200 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs200876773 | 201 | V>F | No |
1000Genomes TOPMed |
|
| rs200876773 | 201 | V>I | No |
1000Genomes TOPMed |
|
| rs1247605228 | 205 | Q>R | No |
TOPMed gnomAD |
|
| rs755605335 | 206 | S>R | No |
ExAC gnomAD |
|
| rs2060039442 | 206 | S>T | No | TOPMed | |
| rs759259237 | 208 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs759259237 | 208 | Y>N | No |
ExAC TOPMed gnomAD |
|
| COSM3547249 | 209 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774150310 | 209 | Q>L | No |
ExAC gnomAD |
|
| rs1272453055 | 210 | K>M | No | gnomAD | |
| rs2145816612 | 211 | I>V | No | Ensembl | |
| rs1231461916 | 212 | A>T | No |
TOPMed gnomAD |
|
| rs2060038867 | 212 | A>V | No | Ensembl | |
| rs373492045 | 213 | E>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs2060038591 | 213 | E>D | No | TOPMed | |
|
COSM3423692 COSM3423691 rs373492045 |
213 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs113022678 | 215 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1296399527 | 216 | T>A | No | gnomAD | |
| rs1452836231 | 216 | T>N | No | gnomAD | |
| rs140516654 | 217 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs140516654 | 217 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs796338768 | 218 | G>V | No | gnomAD | |
| rs776145871 | 220 | L>I | No |
ExAC gnomAD |
|
| rs746249646 | 221 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs779373960 | 223 | H>Q | No |
ExAC gnomAD |
|
| rs2060037722 | 223 | H>Y | No | TOPMed | |
| rs112131818 | 224 | L>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs112131818 | 224 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777396145 | 225 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1438514680 | 226 | V>F | No |
TOPMed gnomAD |
|
| rs1345979268 | 227 | S>C | No | TOPMed | |
| rs2059992166 | 227 | S>P | No |
TOPMed gnomAD |
|
| rs1289610603 | 228 | N>S | No | gnomAD | |
| rs1226077867 | 229 | Q>R | No | gnomAD | |
| rs934300586 | 234 | Y>N | No |
TOPMed gnomAD |
|
| rs764442046 | 235 | A>S | No |
ExAC gnomAD |
|
| rs764442046 | 235 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
|
rs761017231 COSM3547246 COSM3547245 |
236 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2059991519 | 239 | I>M | No | gnomAD | |
| rs767691916 | 240 | N>S | No |
ExAC gnomAD |
|
| TCGA novel | 244 | L>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2059991237 | 246 | A>V | No | gnomAD | |
| TCGA novel | 247 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774923913 | 250 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs771280042 | 251 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs763326281 | 251 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs745858689 | 254 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1315169588 | 254 | M>L | No | gnomAD | |
| rs778989327 | 255 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs757118736 | 256 | N>D | No |
ExAC gnomAD |
|
| rs748974257 | 256 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1369276904 | 259 | A>V | No | gnomAD | |
| rs1368652634 | 261 | K>R | No | gnomAD | |
| rs372415378 | 263 | L>F | No | ESP | |
| rs2145796729 | 264 | R>Q | No | Ensembl | |
|
COSM213406 rs1166553619 |
264 | R>W | breast [Cosmic] | No |
cosmic curated gnomAD |
| rs2059932443 | 266 | I>T | No | Ensembl | |
| rs1319547135 | 272 | I>L | No | TOPMed | |
| rs1319547135 | 272 | I>V | No | TOPMed | |
| rs2059848939 | 273 | R>* | No | TOPMed | |
| rs547937043 | 273 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs547937043 | 273 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs547937043 | 273 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1478278264 | 276 | R>C | No |
TOPMed gnomAD |
|
|
COSM4099012 COSM4099013 rs562401369 |
276 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs549948182 | 278 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1432422732 | 279 | K>E | No | gnomAD | |
| rs746012581 | 279 | K>I | No | Ensembl | |
| rs1248875744 | 281 | E>G | No | gnomAD | |
| TCGA novel | 283 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM70597 | 284 | H>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1474639500 | 284 | H>R | No | Ensembl | |
|
COSM1027455 COSM4874107 |
286 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3841185 COSM3841184 |
290 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1201135584 | 290 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs750445911 | 290 | Q>R | No |
ExAC gnomAD |
|
| rs1467354568 | 291 | V>I | No | TOPMed | |
| rs1470323507 | 293 | T>I | No | Ensembl | |
| rs765755600 | 297 | L>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 299 | E>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1366465262 | 300 | R>K | No | gnomAD | |
| rs779210691 | 302 | M>T | No | Ensembl | |
| rs2059847014 | 304 | K>T | No | TOPMed | |
| rs754339994 | 306 | D>E | No |
ExAC gnomAD |
|
| rs1316972840 | 308 | N>D | No | gnomAD | |
| rs1414591058 | 308 | N>S | No | gnomAD | |
|
COSM4919398 COSM4919397 |
313 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1164123350 | 313 | R>S | No | gnomAD | |
| rs1057105473 | 314 | D>G | No |
TOPMed gnomAD |
|
|
COSM4782912 COSM1412178 |
314 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs370689922 | 315 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1310701259 | 320 | R>G | No | TOPMed | |
| rs1310701259 | 320 | R>W | No | TOPMed | |
| rs1600832673 | 322 | I>M | No | TOPMed | |
| rs754367115 | 322 | I>T | No |
ExAC gnomAD |
|
|
rs756447733 COSM4099010 COSM4099011 |
326 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs947139087 | 327 | E>K | No |
TOPMed gnomAD |
|
|
COSM4824390 COSM4824389 |
329 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs537485260 | 330 | P>L | No |
1000Genomes ExAC |
|
|
COSM6160076 COSM6160077 |
331 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1459534143 | 332 | N>S | No |
TOPMed gnomAD |
|
| rs751231878 | 333 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs1568755229 | 334 | P>A | No | Ensembl | |
| rs2059835533 | 335 | G>E | No | Ensembl | |
|
COSM5134558 COSM5134557 |
335 | G>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1834417043 | 336 | S>N | No |
TOPMed gnomAD |
|
| rs924016543 | 339 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs762523733 | 341 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1568755136 | 343 | A>P | No | Ensembl | |
| rs773032196 | 344 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1353771224 | 345 | Y>S | No | gnomAD | |
| rs769717007 | 348 | D>Y | No |
ExAC gnomAD |
|
| TCGA novel | 351 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 354 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776370647 | 355 | T>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761620325 | 355 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs761620325 | 355 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs2059834614 | 355 | T>S | No | TOPMed | |
| rs1213077138 | 362 | M>T | No | gnomAD | |
| rs2059817155 | 365 | T>I | No | Ensembl | |
| TCGA novel | 368 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4099008 COSM4099009 |
369 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs140527444 | 375 | D>E | No |
ESP ExAC gnomAD |
|
| rs899980540 | 375 | D>N | No |
TOPMed gnomAD |
|
| rs2059816357 | 379 | Y>S | No | Ensembl | |
| rs2059815768 | 380 | L>* | No | TOPMed | |
| rs1367660033 | 380 | L>F | No | gnomAD | |
| rs2059815953 | 380 | L>M | No | TOPMed | |
| rs781764033 | 381 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs755402406 | 382 | K>E | No |
ExAC gnomAD |
|
| TCGA novel | 384 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1401166744 | 385 | Q>* | No | Ensembl | |
| rs369112036 | 387 | T>S | No |
ESP TOPMed |
|
| rs750012831 | 389 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1448574516 | 390 | R>Q | No | Ensembl | |
| rs748175627 | 393 | L>S | No |
ExAC gnomAD |
|
|
COSM6093481 COSM6093480 |
394 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1336045296 | 394 | E>K | No |
TOPMed gnomAD |
|
| rs1427567051 | 396 | S>G | No |
TOPMed gnomAD |
|
| rs1341983440 | 397 | S>R | No | TOPMed | |
|
COSM4099004 rs769240700 COSM4099005 |
398 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1221194910 | 399 | E>K | No | TOPMed | |
| rs768485829 | 403 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2059809323 | 405 | P>S | No | Ensembl | |
| TCGA novel | 407 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1183123716 | 408 | R>C | No | gnomAD | |
| rs372418988 | 408 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs959809295 | 409 | S>G | No |
TOPMed gnomAD |
|
| rs2145776040 | 410 | A>T | No | Ensembl | |
| rs1483950683 | 411 | I>V | No | gnomAD | |
| rs553031271 | 414 | T>N | No | Ensembl | |
| TCGA novel | 417 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1314053062 COSM6160078 COSM6160079 COSM1533635 |
418 | C>R | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs1322559248 | 421 | L>M | No |
TOPMed gnomAD |
|
| rs1314893915 | 422 | Q>* | No | gnomAD | |
| rs1314893915 | 422 | Q>K | No | gnomAD | |
| rs139479585 | 422 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 423 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770422587 | 425 | E>A | No | TOPMed | |
| rs770422587 | 425 | E>V | No | TOPMed | |
| rs1216332604 | 426 | L>P | No |
TOPMed gnomAD |
|
| rs2059806938 | 427 | P>S | No | TOPMed | |
| rs1016412832 | 431 | R>C | No |
TOPMed gnomAD |
|
|
COSM189007 rs776816963 COSM4292685 |
431 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed |
| rs776816963 | 431 | R>L | No |
ExAC TOPMed |
|
| rs2059776410 | 432 | N>S | No | TOPMed | |
| rs1600826991 | 434 | Y>S | No | Ensembl | |
| rs1306672417 | 436 | P>A | No |
TOPMed gnomAD |
|
| rs761294008 | 436 | P>L | No | ExAC | |
| rs1306672417 | 436 | P>S | No |
TOPMed gnomAD |
|
| rs1367829976 | 437 | M>I | No | gnomAD | |
| rs1212005445 | 437 | M>L | No |
TOPMed gnomAD |
|
| rs1459399300 | 441 | H>R | No | gnomAD | |
| rs1159266919 | 442 | D>E | No | gnomAD | |
| rs2059774901 | 443 | R>* | No | Ensembl | |
|
COSM6093483 COSM6093482 |
443 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1013517659 | 443 | R>Q | No | TOPMed | |
| rs2059774641 | 444 | A>V | No | gnomAD | |
|
COSM1307473 COSM4811436 |
446 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs894699586 | 448 | L>F | No | Ensembl | |
| rs1568751627 | 450 | G>A | No | gnomAD | |
| TCGA novel | 453 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2059773886 | 454 | Q>H | No | TOPMed | |
| rs1362014561 | 460 | W>L | No | gnomAD | |
| TCGA novel | 463 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1180800694 | 464 | R>S | No | gnomAD | |
| COSM280777 | 465 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 466 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2145771853 | 466 | T>S | No | Ensembl | |
| rs772557268 | 467 | A>T | No |
ExAC gnomAD |
|
| rs1192054300 | 471 | N>H | No | gnomAD | |
| rs2059725396 | 474 | M>I | No |
TOPMed gnomAD |
|
| rs1317041559 | 474 | M>L | No | gnomAD | |
| rs2059725524 | 474 | M>T | No |
TOPMed gnomAD |
|
| TCGA novel | 475 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747557564 | 477 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs575255605 | 478 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs575255605 | 478 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1433907729 | 479 | E>Q | No | gnomAD | |
| rs201222160 | 482 | T>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 483 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1394806445 | 487 | S>C | No |
TOPMed gnomAD |
|
| rs747312721 | 489 | P>A | No | gnomAD | |
| TCGA novel | 489 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs574674005 | 490 | N>K | No |
ExAC TOPMed gnomAD |
|
| COSM3547238 | 491 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1255685308 | 494 | Q>E | No |
TOPMed gnomAD |
|
| rs752677688 | 494 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2059722939 | 495 | F>L | No | Ensembl | |
| rs767601980 | 496 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1292758470 | 496 | K>R | No |
TOPMed gnomAD |
|
| rs2059722707 | 497 | S>G | No | TOPMed | |
| rs1274898861 | 497 | S>N | No | gnomAD | |
| rs2059722293 | 500 | R>C | No |
TOPMed gnomAD |
|
|
rs760009580 COSM5755592 |
500 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1269026869 | 502 | L>V | No | gnomAD | |
| rs766793261 | 503 | S>N | No |
ExAC TOPMed gnomAD |
|
|
rs774822992 COSM3841179 |
503 | S>R | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs763234588 COSM3841179 |
503 | S>R | breast [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs772866712 | 505 | S>C | No |
ExAC gnomAD |
|
| rs1600822641 | 505 | S>P | No | TOPMed | |
| rs747728081 | 508 | L>V | No |
ExAC gnomAD |
|
| rs768190847 | 509 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| COSM4099001 | 511 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 514 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2059720134 | 516 | M>I | No |
TOPMed gnomAD |
|
| rs2059719994 | 517 | S>G | No | Ensembl | |
| rs2059719868 | 517 | S>N | No | gnomAD | |
| rs2059719750 | 520 | D>E | No | Ensembl | |
| TCGA novel | 520 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs993357068 | 521 | F>L | No | TOPMed | |
| rs2059719620 | 521 | F>Y | No | Ensembl | |
| rs538756608 | 522 | Q>H | No | 1000Genomes | |
| rs1600822482 | 523 | S>A | No | TOPMed | |
| rs1600822482 | 523 | S>P | No | TOPMed | |
| rs745607349 | 524 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs376458042 | 524 | P>T | No |
ESP ExAC gnomAD |
|
| rs1209903982 | 526 | I>T | No | gnomAD | |
| rs985676110 | 526 | I>V | No |
TOPMed gnomAD |
|
| rs1209732040 | 528 | E>D | No |
TOPMed gnomAD |
|
| TCGA novel | 529 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757057713 | 531 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs778618407 | 531 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1325272111 | 532 | K>R | No | gnomAD | |
| rs748385528 | 533 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs778560733 | 536 | E>K | No | gnomAD | |
| rs2059712556 | 539 | E>Q | No | TOPMed | |
| rs2059712156 | 542 | K>R | No | Ensembl | |
| rs1192428312 | 544 | Q>* | No | gnomAD | |
| rs1376289681 | 545 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2145763718 | 548 | R>Q | No | Ensembl | |
| rs2059711422 | 550 | C>R | No | Ensembl | |
| rs2059711303 | 550 | C>S | No | TOPMed | |
| rs1485948572 | 551 | E>D | No |
TOPMed gnomAD |
|
| rs954941889 | 553 | S>N | No | TOPMed | |
| rs1287203765 | 556 | R>* | No |
TOPMed gnomAD |
|
| rs780571140 | 556 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1280736106 | 559 | G>E | No | gnomAD | |
| rs750919846 | 561 | R>H | No |
ExAC gnomAD |
|
|
COSM1205204 rs765582782 |
562 | R>* | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
rs372963438 COSM4644287 |
562 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2059709710 | 563 | R>K | No | Ensembl | |
| TCGA novel | 565 | E>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1568748031 | 565 | E>K | No | Ensembl | |
| rs750963456 | 566 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1295726035 | 566 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2059704422 | 567 | F>Y | No | gnomAD | |
| rs1454354899 | 571 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1568747792 | 571 | R>W | No | TOPMed | |
| rs1375161044 | 576 | H>Y | No | gnomAD | |
| rs1704878908 | 577 | K>E | No |
TOPMed gnomAD |
|
| rs779413222 | 578 | V>I | No |
ExAC gnomAD |
|
| rs779413222 | 578 | V>L | No |
ExAC gnomAD |
|
| rs1322513261 | 580 | H>D | No |
TOPMed gnomAD |
|
| rs757578934 | 581 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs2059703370 | 582 | G>D | No | Ensembl | |
| rs1355945041 | 584 | L>S | No | gnomAD | |
| rs985623713 | 585 | D>G | No | gnomAD | |
| rs756717915 | 585 | D>H | No | Ensembl | |
| rs756717915 | 585 | D>N | No | Ensembl | |
| rs764520008 | 586 | D>A | No |
ExAC gnomAD |
|
| rs754184468 | 586 | D>N | No |
ExAC gnomAD |
|
| rs760450272 | 587 | N>H | No |
ExAC gnomAD |
|
| rs533065163 | 587 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs752380720 | 587 | N>S | No |
ExAC gnomAD |
|
| rs1177591911 | 588 | P>T | No |
TOPMed gnomAD |
|
| rs1355234621 | 589 | Q>H | No | Ensembl | |
| rs1481727918 | 589 | Q>K | No | gnomAD | |
| TCGA novel | 590 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1369415304 | 593 | T>A | No | TOPMed | |
| rs1389344568 | 593 | T>I | No |
TOPMed gnomAD |
|
| rs1600820824 | 598 | Q>K | No | Ensembl | |
| rs770851300 | 599 | E>* | No |
ExAC gnomAD |
|
| COSM4099000 | 599 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 600 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1565752 | 600 | K>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1338454165 | 602 | P>A | No | Ensembl | |
| rs2059681429 | 603 | V>I | No | gnomAD | |
| rs2059681369 | 604 | A>V | No | TOPMed | |
| rs2059681205 | 609 | I>T | No | Ensembl | |
| rs371778780 | 609 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs773121104 | 614 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs2145758703 | 615 | C>F | No | Ensembl | |
| rs765084505 | 616 | P>L | No |
ExAC gnomAD |
|
| rs1199471389 | 617 | H>N | No | Ensembl | |
| rs761534514 | 618 | M>L | No |
ExAC gnomAD |
|
| rs2059680337 | 622 | S>N | No | TOPMed | |
| rs2059680255 | 625 | K>Q | No | TOPMed | |
| rs368066811 | 627 | N>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1327301991 | 627 | N>S | No | gnomAD | |
| rs1379341667 | 630 | V>A | No |
TOPMed gnomAD |
|
| rs1193707713 | 631 | L>W | No | gnomAD | |
| COSM4922899 | 636 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1244490524 | 638 | L>P | No | gnomAD | |
| rs2059639090 | 639 | Y>S | No | TOPMed | |
| rs1749400007 | 642 | D>G | No |
TOPMed gnomAD |
|
| rs2059639028 | 642 | D>H | No | gnomAD | |
| rs777509765 | 643 | E>K | No |
ExAC gnomAD |
|
| rs969548862 | 644 | T>I | No |
TOPMed gnomAD |
|
| rs969548862 | 644 | T>N | No |
TOPMed gnomAD |
|
| rs2059638637 | 647 | F>V | No | Ensembl | |
|
COSM1027453 rs763315321 |
649 | A>T | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1016720830 | 650 | P>R | No |
TOPMed gnomAD |
|
| rs999596170 | 655 | Y>F | No |
TOPMed gnomAD |
|
| rs2059608285 | 657 | I>T | No | TOPMed | |
| rs2059608427 | 657 | I>V | No | Ensembl | |
| rs1233759030 | 659 | I>T | No | TOPMed | |
| rs759832161 | 659 | I>V | No |
ExAC gnomAD |
|
| rs2257495 | 660 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1181943101 | 661 | G>S | No |
TOPMed gnomAD |
|
| rs868613199 | 669 | D>G | No | Ensembl | |
| rs748101681 | 670 | M>T | No |
ExAC gnomAD |
|
| rs144087106 | 670 | M>V | No |
ESP ExAC gnomAD |
|
| rs1218510410 | 671 | S>F | No | gnomAD | |
| TCGA novel | 671 | S>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs946895887 | 672 | S>T | No | TOPMed | |
| rs2059606977 | 675 | T>I | No | gnomAD | |
| rs1295466207 | 676 | K>R | No | gnomAD | |
| rs2059606491 | 683 | L>P | No | gnomAD | |
| rs1165022748 | 684 | S>N | No |
TOPMed gnomAD |
|
| rs1442483526 | 685 | M>I | No | gnomAD | |
| rs1394259728 | 687 | M>L | No | TOPMed | |
| rs2059606265 | 687 | M>T | No | Ensembl | |
| rs2059606195 | 688 | K>Q | No | TOPMed | |
| rs756148777 | 690 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
|
COSM189006 rs764140896 |
690 | R>W | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
rs34630674 VAR_048928 |
695 | E>D | No |
UniProt Ensembl dbSNP |
|
| rs1376912956 | 695 | E>K | No | gnomAD | |
| rs1177941517 | 696 | N>D | No |
TOPMed gnomAD |
|
| rs1177941517 | 696 | N>H | No |
TOPMed gnomAD |
|
| rs370400358 | 697 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs759921910 | 697 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs146276812 | 698 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1338038536 | 699 | I>S | No |
TOPMed gnomAD |
|
| rs1469286549 | 700 | P>T | No | gnomAD | |
| rs202085930 | 701 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1202077385 | 703 | P>A | No |
TOPMed gnomAD |
|
| rs1568740886 | 704 | P>S | No | TOPMed | |
| rs141818894 | 705 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs776733201 | 705 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs776733201 | 705 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1461893840 | 706 | I>L | No | Ensembl | |
| rs2145744603 | 706 | I>M | No | Ensembl | |
| rs921289345 | 706 | I>N | No | gnomAD | |
| rs921289345 | 706 | I>T | No | gnomAD | |
| rs1371357747 | 708 | K>E | No | Ensembl | |
| rs1371357747 | 708 | K>Q | No | Ensembl | |
| rs1600809795 | 708 | K>R | No | Ensembl | |
| rs746916401 | 709 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs1456343092 | 709 | E>D | No | gnomAD | |
| rs780047843 | 709 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs746916401 | 709 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs780047843 | 709 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs1159602575 | 711 | S>G | No |
TOPMed gnomAD |
|
| rs36000025 | 712 | S>N | No | Ensembl | |
| rs771891555 | 712 | S>R | No |
ExAC gnomAD |
|
| rs2059603102 | 717 | Y>C | No |
TOPMed gnomAD |
|
| rs777989540 | 719 | Y>* | No |
ExAC gnomAD |
|
| rs2059603020 | 719 | Y>C | No | TOPMed | |
| rs1168397907 | 721 | G>R | No |
TOPMed gnomAD |
1 associated diseases with Q96JJ3
[MIM: 606893]: Vascular malformation, primary intraosseous (VMPI)
An autosomal recessive, rare malformation characterized by non-neoplastic severe expansions of blood vessels, usually seen in the vertebral column and in the skull. The most commonly affected bones in the skull are the mandible and the maxilla, and life-threatening bleeding after a simple tooth extraction is frequently observed. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal recessive, rare malformation characterized by non-neoplastic severe expansions of blood vessels, usually seen in the vertebral column and in the skull. The most commonly affected bones in the skull are the mandible and the maxilla, and life-threatening bleeding after a simple tooth extraction is frequently observed. . Note=The disease is caused by variants affecting the gene represented in this entry.
2 GO annotations of cellular component
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| SH3 domain binding | Binding to a SH3 domain (Src homology 3) of a protein, small protein modules containing approximately 50 amino acid residues found in a great variety of intracellular or membrane-associated proteins. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| cell chemotaxis | The directed movement of a motile cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| cell motility | Any process involved in the controlled self-propelled movement of a cell that results in translocation of the cell from one place to another. |
| phagocytosis | A vesicle-mediated transport process that results in the engulfment of external particulate material by phagocytes and their delivery to the lysosome. The particles are initially contained within phagocytic vacuoles (phagosomes), which then fuse with primary lysosomes to effect digestion of the particles. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A4FUD6 | ELMO2 | Engulfment and cell motility protein 2 | Bos taurus (Bovine) | SS |
| Q8IZ81 | ELMOD2 | ELMO domain-containing protein 2 | Homo sapiens (Human) | PR |
| Q92556 | ELMO1 | Engulfment and cell motility protein 1 | Homo sapiens (Human) | EV |
| Q96BJ8 | ELMO3 | Engulfment and cell motility protein 3 | Homo sapiens (Human) | SS |
| Q8BPU7 | Elmo1 | Engulfment and cell motility protein 1 | Mus musculus (Mouse) | SS |
| Q8BYZ7 | Elmo3 | Engulfment and cell motility protein 3 | Mus musculus (Mouse) | SS |
| Q8BHL5 | Elmo2 | Engulfment and cell motility protein 2 | Mus musculus (Mouse) | SS |
| Q499U2 | Elmo3 | Engulfment and cell motility protein 3 | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPPPSDIVKV | AIEWPGANAQ | LLEIDQKRPL | ASIIKEVCDG | WSLPNPEYYT | LRYADGPQLY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ITEQTRSDIK | NGTILQLAIS | PSRAARQLME | RTQSSNMETR | LDAMKELAKL | SADVTFATEF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| INMDGIIVLT | RLVESGTKLL | SHYSEMLAFT | LTAFLELMDH | GIVSWDMVSI | TFIKQIAGYV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SQPMVDVSIL | QRSLAILESM | VLNSQSLYQK | IAEEITVGQL | ISHLQVSNQE | IQTYAIALIN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ALFLKAPEDK | RQDMANAFAQ | KHLRSIILNH | VIRGNRPIKT | EMAHQLYVLQ | VLTFNLLEER |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MMTKMDPNDQ | AQRDIIFELR | RIAFDAESDP | SNAPGSGTEK | RKAMYTKDYK | MLGFTNHINP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| AMDFTQTPPG | MLALDNMLYL | AKVHQDTYIR | IVLENSSRED | KHECPFGRSA | IELTKMLCEI |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LQVGELPNEG | RNDYHPMFFT | HDRAFEELFG | ICIQLLNKTW | KEMRATAEDF | NKVMQVVREQ |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ITRALPSKPN | SLDQFKSKLR | SLSYSEILRL | RQSERMSQDD | FQSPPIVELR | EKIQPEILEL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| IKQQRLNRLC | EGSSFRKIGN | RRRQERFWYC | RLALNHKVLH | YGDLDDNPQG | EVTFESLQEK |
| 610 | 620 | 630 | 640 | 650 | 660 |
| IPVADIKAIV | TGKDCPHMKE | KSALKQNKEV | LELAFSILYD | PDETLNFIAP | NKYEYCIWID |
| 670 | 680 | 690 | 700 | 710 | |
| GLSALLGKDM | SSELTKSDLD | TLLSMEMKLR | LLDLENIQIP | EAPPPIPKEP | SSYDFVYHYG |