Q96HU8
Gene name |
DIRAS2 |
Protein name |
GTP-binding protein Di-Ras2 |
Names |
Distinct subgroup of the Ras family member 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:54769 |
EC number |
|
Protein Class |
|
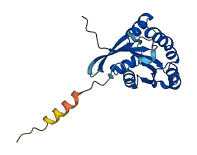
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

3 structures for Q96HU8
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2ERX | X-ray | 165 A | A/B | 5-176 | PDB |
| 6NAZ | X-ray | 308 A | PDB | ||
| AF-Q96HU8-F1 | Predicted | AlphaFoldDB |
124 variants for Q96HU8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs761066904 CA5119084 |
2 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1342076040 CA373834945 |
4 | Q>R | No |
ClinGen gnomAD |
|
|
CA373834924 rs1358140035 |
7 | D>H | No |
ClinGen TOPMed |
|
|
CA373834910 rs1401702721 |
8 | Y>* | No |
ClinGen TOPMed |
|
|
rs768090981 CA5119082 |
9 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA373834905 rs1435305252 |
10 | V>M | No |
ClinGen gnomAD |
|
|
rs1322302173 CA373834892 |
12 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA373834893 rs1322302173 |
12 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
rs930935634 CA195798169 |
17 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
CA195798166 rs142167721 |
18 | V>A | No |
ClinGen ESP |
|
|
CA373834838 rs1163700163 |
20 | K>N | No |
ClinGen gnomAD |
|
|
rs772371364 CA5119074 |
22 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA373834795 rs1587718331 |
27 | F>V | No |
ClinGen Ensembl |
|
|
CA5119071 rs746879754 |
31 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5119069 rs372660715 |
33 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5119070 COSM268220 rs779203948 |
33 | R>W | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
| TCGA novel | 34 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA195798153 rs144995318 |
35 | S>N | No |
ClinGen ESP TOPMed |
|
|
rs756237190 CA5119066 |
38 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM295374 CA5119065 rs753119264 |
39 | T>M | Variant assessed as Somatic; 0.0 impact. large_intestine breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA373834705 rs1276201205 |
41 | E>* | No |
ClinGen gnomAD |
|
|
CA5119062 rs755544272 |
43 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs762495315 CA5119059 |
48 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA5119057 rs764964710 |
53 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA373834579 rs147357334 |
58 | Q>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA373834569 rs1587718273 |
60 | T>P | No |
ClinGen Ensembl |
|
| TCGA novel | 61 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA373834552 rs1564018009 |
62 | T>M | No |
ClinGen Ensembl |
|
|
rs1158778730 CA373834550 |
63 | T>A | No |
ClinGen TOPMed |
|
|
rs775534494 CA5119051 |
63 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA373834547 rs775534494 |
63 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 72 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5119048 rs552701958 |
73 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs552701958 CA195798131 |
73 | R>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs552701958 CA195798132 |
73 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA195798127 rs74418559 |
75 | S>A | No |
ClinGen Ensembl |
|
|
rs1374003225 CA373834462 |
76 | I>T | No |
ClinGen gnomAD |
|
|
CA195798125 rs71494416 |
77 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs748312187 CA5119046 |
78 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA373834454 rs748312187 |
78 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1229692314 CA373834430 |
81 | A>G | No |
ClinGen gnomAD |
|
|
CA373834418 rs1564017978 |
83 | I>V | No |
ClinGen Ensembl |
|
|
CA5119043 rs141028232 |
88 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1564017963 CA373834378 |
89 | T>S | No |
ClinGen Ensembl |
|
|
rs766903718 CA5119042 |
90 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs758782883 CA5119041 |
91 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1484481314 CA373834322 |
98 | K>Q | No |
ClinGen gnomAD |
|
|
rs943848170 CA195798113 |
100 | I>V | No |
ClinGen Ensembl |
|
|
CA5119036 rs567589045 |
102 | E>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA373834268 rs1433853836 |
105 | C>S | No |
ClinGen gnomAD |
|
|
CA5119033 rs772145505 |
106 | E>G | No |
ClinGen ExAC |
|
|
CA5119034 rs775373035 |
106 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA |
|
rs1229715685 CA373834247 |
108 | K>R | No |
ClinGen gnomAD |
|
|
CA373834238 rs1177576373 |
109 | G>A | No |
ClinGen TOPMed |
|
|
rs1316965681 CA373834230 |
110 | D>E | No |
ClinGen gnomAD |
|
|
rs1226534643 CA373834226 |
111 | V>A | No |
ClinGen gnomAD |
|
|
rs759324657 CA5119032 COSM753909 |
111 | V>L | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1441209552 CA373834211 |
113 | S>I | No |
ClinGen TOPMed |
|
| TCGA novel | 113 | S>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5119030 rs769748046 |
114 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA373834201 rs1386770268 |
115 | P>A | No |
ClinGen gnomAD |
|
|
rs911090810 CA195798099 |
115 | P>R | No |
ClinGen TOPMed |
|
| TCGA novel | 116 | I>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1456276902 CA373834126 |
125 | E>D | No |
ClinGen TOPMed |
|
|
CA5119026 rs747397458 |
126 | S>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747397458 CA373834121 |
126 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5119025 rs780339793 |
126 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs986198864 CA195798091 |
127 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1415069140 CA373834116 |
127 | P>S | No |
ClinGen gnomAD |
|
|
CA373834102 rs1564017915 |
129 | R>H | No |
ClinGen Ensembl |
|
|
rs753583908 CA373834093 |
130 | E>D | No |
ClinGen TOPMed |
|
|
rs1210177521 CA373834094 |
130 | E>V | No |
ClinGen gnomAD |
|
|
rs953609126 CA195798087 |
132 | Q>* | No |
ClinGen Ensembl |
|
|
rs778547186 CA373834062 |
135 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA5119022 rs778547186 |
135 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763857662 CA5119019 |
137 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5119020 rs753355155 |
137 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA373834032 rs1357435129 |
139 | L>F | No |
ClinGen TOPMed |
|
|
rs752544271 CA5119017 |
140 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs76263916 CA5119016 |
141 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1430105570 CA373834020 |
142 | T>A | No |
ClinGen gnomAD |
|
|
rs1390213971 CA373833999 |
144 | K>N | No |
ClinGen gnomAD |
|
|
CA373834001 rs1198374954 |
144 | K>R | No |
ClinGen TOPMed |
|
|
rs759573694 CA5119015 |
146 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA373833970 rs1459468281 |
148 | M>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1256635186 CA373833976 |
148 | M>L | No |
ClinGen TOPMed |
|
|
CA195798076 rs1015133741 |
150 | T>I | No |
ClinGen Ensembl |
|
|
CA373833925 rs1564017883 |
155 | N>S | No |
ClinGen Ensembl |
|
|
CA373833914 rs1198898588 |
156 | H>Q | No |
ClinGen TOPMed |
|
|
rs1458806120 CA373833916 |
156 | H>R | No |
ClinGen TOPMed |
|
|
rs1168480763 CA373833891 |
159 | K>N | No |
ClinGen gnomAD |
|
|
rs769958205 CA5119013 |
161 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs905188763 CA195798069 |
166 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs905188763 CA373833847 |
166 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA373833843 rs1188982569 |
167 | N>H | No |
ClinGen gnomAD |
|
|
rs1489148682 CA373833837 |
167 | N>K | No |
ClinGen gnomAD |
|
| TCGA novel | 167 | N>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs747172290 CA5119009 |
171 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA5119010 rs747172290 |
171 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs374125849 CA5119008 |
171 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA373833814 rs374125849 |
171 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs747172290 CA195798062 |
171 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA373833799 rs1276527017 |
174 | V>M | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 177 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1227806428 CA373833775 |
177 | Q>R | No |
ClinGen gnomAD |
|
|
CA373833765 rs904283931 |
178 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
rs755753351 COSM228508 CA5119007 |
179 | D>N | skin [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs972306292 CA195798055 |
180 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
rs746438687 CA5119006 |
180 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757730938 CA5119005 |
181 | K>* | No |
ClinGen ExAC gnomAD |
|
|
CA5119004 rs757730938 |
181 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1021256467 CA195798051 |
181 | K>R | No |
ClinGen Ensembl |
|
|
CA5119003 rs753408149 |
182 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA373833744 rs1587718057 |
182 | K>R | No |
ClinGen Ensembl |
|
| TCGA novel | 184 | K>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA195798045 rs370332203 |
190 | E>D | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs74349275 CA195798040 |
192 | L>I | No |
ClinGen gnomAD |
|
|
CA373833671 rs74349275 |
192 | L>V | No |
ClinGen gnomAD |
|
|
rs752384572 CA373833655 |
194 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs752384572 CA5118999 |
194 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs1173493031 CA373833647 |
195 | K>N | No |
ClinGen gnomAD |
|
|
CA5118998 rs566281003 |
197 | V>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA373833628 rs1587718025 |
198 | I>T | No |
ClinGen Ensembl |
|
|
rs749734783 CA5118997 |
200 | M>= | No |
ClinGen ExAC gnomAD |
No associated diseases with Q96HU8
1 regional properties for Q96HU8
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 6 - 165 | IPR005225 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
34 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P08642 | HRAS | GTPase HRas | Gallus gallus (Chicken) | SS |
| Q6T310 | RASL11A | Ras-like protein family member 11A | Homo sapiens (Human) | PR |
| Q8IYK8 | REM2 | GTP-binding protein REM 2 | Homo sapiens (Human) | PR |
| P55040 | GEM | GTP-binding protein GEM | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q9BU20 | CPLANE2 | Ciliogenesis and planar polarity effector 2 | Homo sapiens (Human) | PR |
| P11233 | RALA | Ras-related protein Ral-A | Homo sapiens (Human) | PR |
| P62070 | RRAS2 | Ras-related protein R-Ras2 | Homo sapiens (Human) | PR |
| P11234 | RALB | Ras-related protein Ral-B | Homo sapiens (Human) | PR |
| Q99578 | RIT2 | GTP-binding protein Rit2 | Homo sapiens (Human) | PR |
| P01116 | KRAS | GTPase KRas | Homo sapiens (Human) | EV |
| P01112 | HRAS | GTPase HRas | Homo sapiens (Human) | SS |
| Q9JIW9 | Ralb | Ras-related protein Ral-B | Mus musculus (Mouse) | PR |
| Q61411 | Hras | GTPase HRas | Mus musculus (Mouse) | SS |
| P32883 | Kras | GTPase KRas | Mus musculus (Mouse) | SS |
| O08989 | Mras | Ras-related protein M-Ras | Mus musculus (Mouse) | PR |
| Q91Z61 | Diras1 | GTP-binding protein Di-Ras1 | Mus musculus (Mouse) | PR |
| P62071 | Rras2 | Ras-related protein R-Ras2 | Mus musculus (Mouse) | PR |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| Q08AT1 | Rasl12 | Ras-like protein family member 12 | Mus musculus (Mouse) | PR |
| A2A825 | Cplane2 | Ciliogenesis and planar polarity effector 2 | Mus musculus (Mouse) | PR |
| P55041 | Gem | GTP-binding protein GEM | Mus musculus (Mouse) | PR |
| P70425 | Rit2 | GTP-binding protein Rit2 | Mus musculus (Mouse) | PR |
| Q8VEL9 | Rem2 | GTP-binding protein REM 2 | Mus musculus (Mouse) | PR |
| Q5PR73 | Diras2 | GTP-binding protein Di-Ras2 | Mus musculus (Mouse) | PR |
| P36860 | Ralb | Ras-related protein Ral-B | Rattus norvegicus (Rat) | PR |
| P08644 | Kras | GTPase KRas | Rattus norvegicus (Rat) | SS |
| Q9WTY2 | Rem2 | GTP-binding protein REM 2 | Rattus norvegicus (Rat) | PR |
| P20171 | Hras | GTPase HRas | Rattus norvegicus (Rat) | SS |
| Q5BJQ5 | Rit2 | GTP-binding protein Rit2 | Rattus norvegicus (Rat) | PR |
| P97538 | Mras | Ras-related protein M-Ras | Rattus norvegicus (Rat) | PR |
| B7ZTR0 | cplane2 | Ciliogenesis and planar polarity effector 2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| P79737 | nras | GTPase NRas | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A1DZY4 | zgc:110179 | Ras-like protein family member 11A-like | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPEQSNDYRV | AVFGAGGVGK | SSLVLRFVKG | TFRESYIPTV | EDTYRQVISC | DKSICTLQIT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DTTGSHQFPA | MQRLSISKGH | AFILVYSITS | RQSLEELKPI | YEQICEIKGD | VESIPIMLVG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NKCDESPSRE | VQSSEAEALA | RTWKCAFMET | SAKLNHNVKE | LFQELLNLEK | RRTVSLQIDG |
| 190 | |||||
| KKSKQQKRKE | KLKGKCVIM |