Q96G01
Gene name |
BICD1 |
Protein name |
Protein bicaudal D homolog 1 |
Names |
Bic-D 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:636 |
EC number |
|
Protein Class |
|
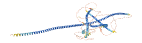
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1-300 (CC1 region) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q96G01
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q96G01-F1 | Predicted | AlphaFoldDB |
881 variants for Q96G01
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1375791160 | 3 | A>G | No | gnomAD | |
| rs1314450126 | 3 | A>S | No |
TOPMed gnomAD |
|
| rs1056581472 | 5 | E>A | No |
TOPMed gnomAD |
|
| rs770500759 | 5 | E>D | No |
ExAC gnomAD |
|
| rs1056581472 | 5 | E>G | No |
TOPMed gnomAD |
|
| rs1176204686 | 5 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1592305287 | 6 | V>G | No | Ensembl | |
| rs770378822 | 8 | Q>E | No |
ExAC gnomAD |
|
| rs200755138 | 8 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs202045108 | 9 | T>K | No | TOPMed | |
| rs202045108 | 9 | T>M | No | TOPMed | |
| rs1404869249 | 9 | T>S | No |
TOPMed gnomAD |
|
| rs1203331100 | 10 | V>G | No | gnomAD | |
| rs764284872 | 11 | D>G | No |
ExAC gnomAD |
|
| rs145280733 | 11 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs377203999 | 12 | H>D | No |
ESP ExAC gnomAD |
|
| rs1430864003 | 13 | Y>* | No | gnomAD | |
| rs1254678328 | 13 | Y>C | No | gnomAD | |
| rs370389517 | 14 | K>E | No |
ESP ExAC gnomAD |
|
| rs1372035952 | 14 | K>N | No | gnomAD | |
| rs926111124 | 14 | K>R | No | Ensembl | |
| rs1941516020 | 15 | T>I | No | TOPMed | |
| rs1387725049 | 15 | T>P | No | TOPMed | |
| rs753716472 | 16 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs754697901 | 17 | I>R | No | ExAC | |
| rs1941517350 | 17 | I>V | No | TOPMed | |
| rs1012800805 | 18 | E>D | No |
TOPMed gnomAD |
|
| rs137871150 | 19 | R>T | No |
ESP ExAC gnomAD |
|
|
COSM1361244 rs1941518234 |
20 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs1941518533 | 21 | T>A | No |
TOPMed gnomAD |
|
| rs200843440 | 22 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs202153353 | 25 | T>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs565110613 | 25 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs565110613 | 25 | T>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM938793 | 26 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs888011675 | 26 | E>K | No | TOPMed | |
| rs1348950786 | 27 | T>I | No | gnomAD | |
| rs1941520030 | 28 | T>A | No | Ensembl | |
| rs201825812 | 29 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1272562402 | 30 | E>Q | No | TOPMed | |
| COSM4920361 | 38 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1227816106 | 38 | G>R | No |
TOPMed gnomAD |
|
| rs1592305691 | 40 | V>A | No | gnomAD | |
| rs1592305691 | 40 | V>G | No | gnomAD | |
| rs200965468 | 41 | V>A | No | 1000Genomes | |
| rs200965468 | 41 | V>G | No | 1000Genomes | |
| rs770082040 | 43 | E>G | No |
ExAC gnomAD |
|
| rs1446127816 | 44 | E>V | No | Ensembl | |
| rs1565517912 | 45 | K>R | No | Ensembl | |
| rs1365363427 | 46 | L>M | No | gnomAD | |
| rs1365363427 | 46 | L>V | No | gnomAD | |
| rs1046103078 | 48 | L>F | No | gnomAD | |
| rs1046103078 | 48 | L>V | No | gnomAD | |
| rs769283505 | 49 | K>I | No |
ExAC TOPMed gnomAD |
|
| rs769283505 | 49 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs1592305818 | 50 | Q>H | No | Ensembl | |
| rs1432982556 | 50 | Q>L | No | gnomAD | |
| rs1328072045 | 51 | Q>R | No |
TOPMed gnomAD |
|
| rs1386460297 | 52 | Y>H | No | gnomAD | |
| rs115927890 | 53 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1319050732 | 54 | E>G | No | gnomAD | |
| rs1167385966 | 57 | A>S | No | Ensembl | |
| rs1260584460 | 57 | A>V | No | gnomAD | |
| rs1327785580 | 58 | E>G | No |
TOPMed gnomAD |
|
| rs961986142 | 58 | E>K | No | TOPMed | |
| rs1327785580 | 58 | E>V | No |
TOPMed gnomAD |
|
| rs1215008217 | 59 | Y>H | No | gnomAD | |
| rs1467728893 | 60 | D>Y | No | gnomAD | |
| rs1415362767 | 61 | S>G | No |
TOPMed gnomAD |
|
|
TCGA novel rs1941527481 |
61 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs567663900 | 61 | S>R | No |
1000Genomes TOPMed gnomAD |
|
| rs199983081 | 63 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs760902488 | 64 | Q>L | No |
ExAC TOPMed gnomAD |
|
| COSM6072460 | 65 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201098113 | 66 | L>V | No | TOPMed | |
| rs1941528256 | 67 | E>K | No | TOPMed | |
| rs1592306028 | 68 | Q>H | No |
TOPMed gnomAD |
|
| rs764953535 | 68 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs1167149614 | 70 | K>R | No | gnomAD | |
| rs955091441 | 72 | A>S | No | TOPMed | |
| rs1945357816 | 72 | A>V | No | Ensembl | |
|
rs1014050024 COSM4876825 |
77 | F>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| COSM4924287 | 78 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs986503549 COSM4041416 |
79 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs199513734 | 80 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs747095378 | 80 | H>Y | No |
ExAC gnomAD |
|
| rs776799556 | 81 | R>Q | No |
ExAC gnomAD |
|
| rs1322401627 | 81 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1279971081 | 83 | V>L | No | gnomAD | |
| rs1248504065 | 84 | A>S | No |
TOPMed gnomAD |
|
| rs1248504065 | 84 | A>T | No |
TOPMed gnomAD |
|
| rs1349762231 | 84 | A>V | No | gnomAD | |
| rs540419761 | 85 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1295076942 | 86 | D>V | No | gnomAD | |
| rs1164857152 | 87 | G>R | No | gnomAD | |
| rs761725856 | 88 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs200761672 | 88 | E>D | No |
TOPMed gnomAD |
|
| rs749951513 | 90 | R>Q | No |
ExAC gnomAD |
|
| COSM4938403 | 91 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1945360209 | 92 | E>D | No | gnomAD | |
| rs1592493219 | 92 | E>G | No | Ensembl | |
| rs777630104 | 94 | L>F | No |
ExAC gnomAD |
|
| rs753565776 | 95 | L>R | No |
ExAC gnomAD |
|
| rs1422604302 | 96 | Q>R | No | gnomAD | |
| rs140970877 | 102 | E>D | No |
1000Genomes TOPMed gnomAD |
|
| COSM6136777 | 102 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1945361276 | 103 | A>T | No | Ensembl | |
| COSM5954978 | 103 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1492892 | 104 | Y>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1945361377 | 104 | Y>S | No | Ensembl | |
| rs753099229 | 105 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1945361562 | 106 | L>M | No | Ensembl | |
| COSM431029 | 106 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1592493340 | 107 | G>E | No | Ensembl | |
| rs1429550798 | 107 | G>R | No | TOPMed | |
| rs1565593717 | 110 | L>V | No | Ensembl | |
| rs1165066664 | 110 | L>W | No | TOPMed | |
| COSM468279 | 111 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1422408285 | 111 | E>D | No | TOPMed | |
| rs1314570582 | 112 | M>I | No |
TOPMed gnomAD |
|
| rs201462520 | 112 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs200651524 | 112 | M>V | No |
ExAC gnomAD |
|
| rs1444064999 | 113 | Q>R | No | gnomAD | |
| COSM3460196 | 115 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs945989848 | 116 | L>M | No |
TOPMed gnomAD |
|
| rs945989848 | 116 | L>V | No |
TOPMed gnomAD |
|
| rs781194681 | 117 | K>N | No |
ExAC gnomAD |
|
| rs1945363349 | 118 | Q>E | No |
TOPMed gnomAD |
|
| rs1945363501 | 120 | R>W | No |
TOPMed gnomAD |
|
| rs746097394 | 122 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs746097394 | 122 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1482900716 | 123 | V>I | No | gnomAD | |
| rs1471080932 | 124 | T>A | No | TOPMed | |
| rs1945364347 | 124 | T>S | No | Ensembl | |
| rs1945364444 | 125 | N>I | No | gnomAD | |
| rs771944171 | 126 | V>A | No |
ExAC gnomAD |
|
| rs528926009 | 126 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199991019 | 129 | E>G | No | Ensembl | |
| rs1945365192 | 130 | N>D | No | Ensembl | |
| rs1945365386 | 131 | E>D | No | TOPMed | |
| rs773234983 | 131 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs765998934 | 135 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs765998934 | 135 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2121554855 | 135 | A>V | No | Ensembl | |
| rs200613741 | 137 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs200613741 | 137 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs759329449 | 138 | Q>* | No |
ExAC gnomAD |
|
| rs764969519 | 138 | Q>P | No |
ExAC gnomAD |
|
| rs148155238 | 139 | D>N | No |
ESP ExAC gnomAD |
|
| rs1332154103 | 140 | L>M | No |
TOPMed gnomAD |
|
| rs758753948 | 140 | L>P | No |
ExAC gnomAD |
|
| TCGA novel | 141 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1016581569 | 141 | K>R | No | TOPMed | |
| COSM4403808 | 143 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201230666 | 146 | M>T | No | Ensembl | |
| rs1353540741 | 146 | M>V | No |
TOPMed gnomAD |
|
| rs1266982815 | 147 | V>A | No |
TOPMed gnomAD |
|
| rs1208783503 | 147 | V>M | No |
TOPMed gnomAD |
|
|
COSM938795 rs996357022 |
148 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs764526696 | 149 | L>P | No |
ExAC gnomAD |
|
| rs762461502 | 149 | L>V | No |
ExAC gnomAD |
|
| COSM4041417 | 151 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs989540206 | 153 | R>G | No |
TOPMed gnomAD |
|
| rs762369260 | 153 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs989540206 | 153 | R>W | No |
TOPMed gnomAD |
|
| rs200263725 | 154 | M>R | No | Ensembl | |
| rs1228877908 | 156 | D>N | No | gnomAD | |
| rs1457751453 | 159 | R>* | No | gnomAD | |
| rs968259805 | 159 | R>L | No |
TOPMed gnomAD |
|
| rs968259805 | 159 | R>Q | No |
TOPMed gnomAD |
|
| rs767993717 | 161 | Y>C | No |
ExAC gnomAD |
|
| rs779950721 | 164 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs756217437 | 164 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs945384185 | 167 | R>Q | No | TOPMed | |
| rs999370711 | 167 | R>W | No |
TOPMed gnomAD |
|
| COSM265432 | 168 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 170 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200303935 | 172 | Y>C | No | Ensembl | |
| rs201545601 | 177 | E>* | No | Ensembl | |
| TCGA novel | 178 | E>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755087506 | 180 | I>T | No |
ExAC gnomAD |
|
| TCGA novel | 181 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 182 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs924563503 | 184 | K>E | No | TOPMed | |
| rs199528142 | 188 | T>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199528142 | 188 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200529716 | 190 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs200529716 | 190 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs781082556 | 201 | H>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 202 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 205 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs199924935 | 205 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1386207177 | 206 | F>L | No |
TOPMed gnomAD |
|
| rs180887157 | 209 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs367637446 COSM4041419 |
210 | T>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs367637446 | 210 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs367637446 | 210 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs375970572 | 210 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 214 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1948194153 | 217 | L>V | No | gnomAD | |
| rs1272258886 | 219 | D>G | No |
TOPMed gnomAD |
|
| rs199903813 | 220 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1948194541 | 222 | R>* | No | TOPMed | |
| rs907706694 | 222 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs201006198 | 225 | E>Q | No |
TOPMed gnomAD |
|
| rs202126519 | 226 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1180260676 | 228 | E>G | No | gnomAD | |
| rs1450382734 | 230 | Q>L | No | gnomAD | |
| rs1188298106 | 231 | L>M | No | gnomAD | |
| rs766781409 | 231 | L>P | No |
ExAC gnomAD |
|
| TCGA novel | 233 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1416543587 | 234 | A>S | No | gnomAD | |
| rs1948196198 | 236 | E>D | No | Ensembl | |
| COSM938796 | 236 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs139179793 | 236 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1948196286 | 238 | L>S | No | Ensembl | |
| rs1948196374 | 239 | K>R | No | Ensembl | |
| rs377261678 | 240 | N>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs377261678 | 240 | N>H | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 240 | N>M | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1272669967 | 242 | R>I | No | gnomAD | |
| rs1219504340 | 243 | E>G | No | TOPMed | |
| rs1313173588 | 243 | E>Q | No | gnomAD | |
| rs1433850630 | 244 | Q>R | No | gnomAD | |
| rs202121002 | 247 | N>D | No | TOPMed | |
| rs1264580521 | 249 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs758632838 | 249 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs767421423 | 250 | K>N | No |
ExAC gnomAD |
|
| TCGA novel | 252 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200338023 | 254 | Q>E | No |
ExAC gnomAD |
|
| rs1040356579 | 254 | Q>R | No |
TOPMed gnomAD |
|
| rs779829813 | 255 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs779829813 | 255 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs749213007 | 257 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs749213007 | 257 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1948199294 | 258 | L>F | No | TOPMed | |
| rs1948199294 | 258 | L>I | No | TOPMed | |
| rs1248184964 | 259 | N>D | No | gnomAD | |
| rs747806024 | 259 | N>S | No |
ExAC gnomAD |
|
| rs771710502 | 260 | D>N | No | ExAC | |
| rs1948200225 | 261 | N>K | No | gnomAD | |
| rs747312377 | 262 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs773624169 | 262 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1948200685 | 263 | I>T | No | TOPMed | |
| rs199877174 | 267 | V>E | No |
ExAC TOPMed gnomAD |
|
| rs199877174 | 267 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs760036729 | 268 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs760036729 | 268 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1477756670 | 270 | L>F | No |
TOPMed gnomAD |
|
| rs145831771 | 271 | K>R | No |
1000Genomes ExAC gnomAD |
|
| rs546702617 | 272 | F>L | No |
1000Genomes ExAC gnomAD |
|
| rs763327723 | 272 | F>Y | No |
ExAC gnomAD |
|
| rs764393034 | 273 | A>T | No |
ExAC gnomAD |
|
| rs1351328727 | 273 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs755896932 | 274 | E>D | No |
ExAC gnomAD |
|
| rs1230113779 | 275 | D>A | No |
TOPMed gnomAD |
|
| rs1230113779 | 275 | D>G | No |
TOPMed gnomAD |
|
| rs1365374302 | 275 | D>H | No | gnomAD | |
| rs1288936980 | 276 | G>E | No | gnomAD | |
| COSM4877523 | 276 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1443836967 | 277 | S>N | No |
TOPMed gnomAD |
|
| rs1329872157 | 277 | S>R | No | gnomAD | |
| rs1948202967 | 278 | E>Q | No | TOPMed | |
| rs201123404 | 279 | P>S | No | Ensembl | |
| rs753733709 | 281 | N>H | No |
ExAC gnomAD |
|
| rs199601826 | 281 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1222728786 | 282 | D>G | No | gnomAD | |
| rs147476635 | 283 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1948203455 | 283 | D>G | No | Ensembl | |
| COSM6072458 | 287 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs374149547 | 287 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| COSM938797 | 287 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1565657695 | 290 | H>R | No | TOPMed | |
|
rs777372640 COSM1511999 |
290 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs746632766 | 292 | P>R | No |
ExAC gnomAD |
|
| rs771218516 | 293 | L>V | No |
ExAC gnomAD |
|
| rs1347856000 | 295 | K>E | No | gnomAD | |
| rs770418190 | 296 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1308554117 | 299 | D>N | No | gnomAD | |
| rs199502236 | 300 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1277891597 | 301 | R>P | No | gnomAD | |
| rs1277891597 | 301 | R>Q | No | gnomAD | |
| rs763097210 | 301 | R>W | No |
ExAC gnomAD |
|
| rs1948205447 | 302 | T>S | No | Ensembl | |
| TCGA novel | 309 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762037904 | 309 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1289445563 | 309 | E>K | No | gnomAD | |
| rs771769631 | 310 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs771769631 | 310 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs1253152252 | 312 | N>D | No |
TOPMed gnomAD |
|
| rs753665015 | 312 | N>S | No |
ExAC gnomAD |
|
| rs1472855894 | 313 | P>R | No | gnomAD | |
| rs200645288 | 313 | P>T | No |
TOPMed gnomAD |
|
| rs1948206630 | 314 | V>I | No |
TOPMed gnomAD |
|
| rs752243115 | 316 | D>N | No |
ExAC gnomAD |
|
| rs1284613161 | 316 | D>V | No |
TOPMed gnomAD |
|
| rs555237184 | 318 | F>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs555237184 | 318 | F>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 321 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1342282029 | 322 | N>S | No | gnomAD | |
| rs1283815694 | 326 | I>M | No | gnomAD | |
| rs1338439196 | 326 | I>T | No |
TOPMed gnomAD |
|
| rs1332440422 | 327 | Q>R | No | TOPMed | |
| rs1948207894 | 328 | K>T | No | Ensembl | |
| rs1948207986 | 331 | Q>* | No |
TOPMed gnomAD |
|
| rs1469429616 | 331 | Q>P | No |
TOPMed gnomAD |
|
| rs1275849281 | 334 | M>I | No | gnomAD | |
| rs375989586 | 334 | M>L | No |
ESP TOPMed |
|
| rs1231043402 | 334 | M>T | No | gnomAD | |
| rs1948208628 | 335 | Q>P | No | Ensembl | |
| rs749817151 | 336 | V>I | No |
ExAC gnomAD |
|
| rs1038396187 | 337 | E>Q | No |
TOPMed gnomAD |
|
| rs201669807 | 338 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs183425481 | 338 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs183425481 | 338 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201669807 | 338 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM3368785 | 339 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1163350046 | 340 | K>N | No | TOPMed | |
| rs377071191 | 342 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs201240851 | 344 | L>S | No |
ExAC TOPMed gnomAD |
|
| rs773452780 | 345 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs1303911158 | 346 | N>T | No | gnomAD | |
| rs747184772 | 347 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs769589738 | 351 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1250064200 | 353 | Q>R | No | Ensembl | |
| rs1210714600 | 355 | E>G | No |
TOPMed gnomAD |
|
| rs1948811853 | 355 | E>Q | No | TOPMed | |
| rs1948812298 | 356 | H>Y | No | Ensembl | |
| rs1565673233 | 357 | T>A | No | Ensembl | |
| rs200322489 | 360 | A>E | No | Ensembl | |
| TCGA novel | 360 | A>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1565673246 | 360 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs144146678 | 362 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs932373460 | 363 | E>A | No | Ensembl | |
| rs1211383393 | 363 | E>D | No | gnomAD | |
| rs1232544628 | 365 | H>R | No | TOPMed | |
| rs370202079 | 366 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs142187819 | 367 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs142187819 | 367 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs148689974 | 367 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1948815114 | 369 | H>L | No | Ensembl | |
| rs201356744 | 369 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1323777222 | 369 | H>Y | No |
TOPMed gnomAD |
|
| rs199625504 | 370 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs576393370 | 370 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM938798 | 371 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1391832130 | 372 | T>S | No | gnomAD | |
| rs1948816209 | 373 | E>K | No | TOPMed | |
| rs1404722439 | 374 | H>Y | No | gnomAD | |
| rs1160863630 | 375 | V>I | No | TOPMed | |
| rs1948816977 | 376 | N>S | No | Ensembl | |
| rs1363331550 | 376 | N>Y | No | TOPMed | |
| COSM185625 | 377 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1948817100 | 378 | M>L | No |
TOPMed gnomAD |
|
| rs201857634 | 380 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs201857634 | 380 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs771134310 | 382 | Q>R | No |
ExAC gnomAD |
|
| rs1014205883 | 384 | S>G | No | Ensembl | |
| rs1014205883 | 384 | S>R | No | Ensembl | |
| TCGA novel | 385 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1314649986 | 386 | E>A | No | gnomAD | |
| COSM6072457 | 386 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775318566 | 387 | L>P | No |
ExAC gnomAD |
|
| rs749084742 | 388 | K>E | No |
ExAC gnomAD |
|
| COSM6136776 | 388 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763253358 | 391 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1592679544 | 391 | L>P | No | Ensembl | |
| rs763253358 | 391 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs867107052 | 392 | D>N | No | Ensembl | |
|
rs760462735 COSM4041420 |
393 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC TOPMed gnomAD NCI-TCGA Cosmic NCI-TCGA |
| COSM6136775 | 393 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1948819640 | 394 | E>G | No | Ensembl | |
| rs1249842043 | 394 | E>Q | No | gnomAD | |
| TCGA novel | 395 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200717707 | 397 | R>G | No |
TOPMed gnomAD |
|
| rs140303025 | 397 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs200717707 | 397 | R>W | No |
TOPMed gnomAD |
|
| rs1024567413 | 398 | D>E | No |
TOPMed gnomAD |
|
| rs144053152 | 398 | D>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs144053152 | 398 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1410934385 | 399 | S>A | No |
TOPMed gnomAD |
|
| rs1410934385 | 399 | S>P | No |
TOPMed gnomAD |
|
| rs1410934385 | 399 | S>T | No |
TOPMed gnomAD |
|
| rs1370467760 | 400 | G>E | No |
TOPMed gnomAD |
|
| rs368356881 | 400 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1370467760 | 400 | G>V | No |
TOPMed gnomAD |
|
| rs1948821593 | 401 | E>* | No | Ensembl | |
| rs777895773 | 402 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs758760483 | 402 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1565673617 | 403 | A>D | No | Ensembl | |
| rs751623535 | 403 | A>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 404 | H>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757522782 | 404 | H>R | No |
ExAC gnomAD |
|
| TCGA novel | 404 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766903521 | 405 | D>V | No | Ensembl | |
| TCGA novel | 407 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781426726 | 409 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs2136260240 | 410 | I>N | No | Ensembl | |
| rs1948822725 | 410 | I>V | No | TOPMed | |
| rs1384271185 | 411 | N>D | No | Ensembl | |
| rs565049897 | 412 | G>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1948822875 | 412 | G>S | No | Ensembl | |
| rs1948823393 | 415 | I>F | No | TOPMed | |
| rs774804835 | 420 | Y>C | No | Ensembl | |
| rs866814676 | 421 | R>G | No |
TOPMed gnomAD |
|
| rs961929631 | 421 | R>M | No | gnomAD | |
| rs1183727876 | 421 | R>S | No | gnomAD | |
| rs1475598967 | 424 | V>I | No |
TOPMed gnomAD |
|
| rs778506010 | 425 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs2136260429 | 428 | I>T | No | Ensembl | |
| rs748188229 | 429 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1948824457 | 436 | A>S | No | TOPMed | |
| rs760233757 | 437 | L>S | No |
ExAC TOPMed gnomAD |
|
| rs112294543 | 438 | K>* | No | TOPMed | |
| rs112294543 | 438 | K>E | No | TOPMed | |
| COSM4041421 | 439 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1170284368 | 439 | E>Q | No | gnomAD | |
| rs776365092 | 443 | K>N | No |
ExAC gnomAD |
|
| TCGA novel | 443 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1948825152 | 444 | S>F | No | Ensembl | |
| rs1302974983 | 446 | E>D | No | gnomAD | |
| rs1948825241 | 446 | E>Q | No | TOPMed | |
| rs760224331 | 450 | D>Y | No | Ensembl | |
| rs925652150 | 452 | K>T | No | Ensembl | |
| rs867408250 | 453 | A>T | No | TOPMed | |
| rs1449945830 | 453 | A>V | No | gnomAD | |
| rs199959255 | 454 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs199959255 | 454 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs763572118 | 456 | E>D | No |
ExAC gnomAD |
|
| rs935771046 | 459 | I>L | No |
TOPMed gnomAD |
|
| rs2136260729 | 461 | M>V | No | Ensembl | |
| rs764560534 | 462 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1182305653 | 462 | Y>D | No |
TOPMed gnomAD |
|
|
rs1212294612 COSM3460197 |
464 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1948826684 | 466 | V>M | No | Ensembl | |
| rs201221854 | 467 | T>R | No |
ExAC gnomAD |
|
| rs1363034207 | 468 | S>G | No | TOPMed | |
| rs202173166 | 468 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1250141557 | 471 | K>N | No | gnomAD | |
| rs1450479522 | 473 | T>I | No |
TOPMed gnomAD |
|
| rs1186362925 | 474 | K>R | No | gnomAD | |
| rs750663918 | 475 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs2136260880 | 475 | E>G | No | Ensembl | |
| rs781192681 | 475 | E>K | No |
ExAC TOPMed |
|
| rs370854157 | 476 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs778829618 | 480 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1948827811 | 481 | A>T | No | Ensembl | |
| rs200340826 | 481 | A>V | No | Ensembl | |
| rs1948828165 | 483 | M>I | No | Ensembl | |
| rs1427914231 | 483 | M>V | No |
TOPMed gnomAD |
|
| COSM1705359 | 484 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1948828319 | 485 | K>M | No | TOPMed | |
| rs1467776742 | 486 | E>D | No |
TOPMed gnomAD |
|
| rs146443241 | 486 | E>G | No |
ESP ExAC |
|
| rs191137869 | 488 | Q>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs191137869 | 488 | Q>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs191137869 | 488 | Q>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1388995673 | 490 | M>I | No | gnomAD | |
| rs1948828911 | 491 | T>I | No |
TOPMed gnomAD |
|
| rs1948829069 | 493 | I>R | No | gnomAD | |
| rs1948828991 | 493 | I>V | No | TOPMed | |
| rs1173200986 | 495 | N>D | No | TOPMed | |
| rs116296656 | 495 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs116296656 | 495 | N>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200359075 | 496 | E>A | No | Ensembl | |
| rs200359075 | 496 | E>G | No | Ensembl | |
| rs141919751 | 496 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 497 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1219688425 | 497 | N>K | No | gnomAD | |
| rs150074704 | 499 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs538925243 | 501 | L>P | No |
1000Genomes ExAC gnomAD |
|
| rs769317702 | 502 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs769317702 | 502 | N>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 503 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777926895 | 503 | T>M | No |
TOPMed gnomAD |
|
| rs1948830443 | 504 | A>D | No | TOPMed | |
| rs1948830530 | 506 | D>E | No | Ensembl | |
| rs763271425 | 507 | E>G | No |
ExAC gnomAD |
|
| rs1565674150 | 507 | E>K | No | Ensembl | |
| rs1308897925 | 509 | V>G | No | TOPMed | |
| rs2136261285 | 510 | T>A | No | Ensembl | |
| rs752075587 | 515 | L>S | No |
ExAC gnomAD |
|
| rs1948831111 | 517 | Q>R | No | Ensembl | |
| rs1005676624 | 520 | H>Y | No | gnomAD | |
| rs1479161722 | 521 | H>L | No |
TOPMed gnomAD |
|
| rs1479161722 | 521 | H>P | No |
TOPMed gnomAD |
|
| rs1412211101 | 523 | C>S | No | gnomAD | |
| rs767703880 | 525 | C>R | No |
ExAC gnomAD |
|
| rs1351344999 | 525 | C>W | No | gnomAD | |
| rs1317743602 | 526 | N>D | No |
TOPMed gnomAD |
|
| rs1948832015 | 528 | E>V | No | TOPMed | |
| rs1948832094 | 529 | T>A | No | TOPMed | |
| rs1294408537 | 531 | N>K | No |
TOPMed gnomAD |
|
| rs1948832324 | 532 | R>S | No | TOPMed | |
| rs1948832485 | 539 | R>K | No | TOPMed | |
| TCGA novel | 540 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376363261 | 541 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs376363261 | 541 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1948832791 | 542 | R>K | No | TOPMed | |
| rs754053760 | 543 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs754053760 | 543 | V>D | No |
ExAC TOPMed gnomAD |
|
|
rs1223369923 COSM4041422 |
545 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs777649808 | 545 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs777649808 | 545 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs777649808 | 545 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1223761034 | 546 | S>N | No | gnomAD | |
| rs1331354334 | 547 | G>S | No | gnomAD | |
| rs780811727 | 550 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs954277993 | 551 | G>R | No | Ensembl | |
| rs201212053 | 552 | P>L | No | Ensembl | |
| rs1232904133 | 552 | P>T | No | gnomAD | |
| rs201635417 | 553 | D>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs201635417 | 553 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs775034841 | 554 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs748883894 | 555 | P>A | No |
ExAC gnomAD |
|
| rs1162752113 | 555 | P>H | No | gnomAD | |
| rs1948834750 | 557 | G>A | No |
TOPMed gnomAD |
|
| rs1431510952 | 557 | G>R | No | gnomAD | |
| rs774694561 | 559 | L>S | No |
ExAC gnomAD |
|
| rs1416434917 | 560 | S>P | No |
TOPMed gnomAD |
|
| rs1441648772 | 561 | P>S | No |
TOPMed gnomAD |
|
| rs200597704 | 562 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs200597704 | 562 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs201571553 | 562 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201571553 | 562 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM1628562 | 564 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754124048 | 566 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs755181806 | 566 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs754124048 | 566 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs199952837 | 567 | G>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs751383007 | 567 | G>D | No |
ExAC gnomAD |
|
| rs199952837 | 567 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs751383007 | 567 | G>V | No |
ExAC gnomAD |
|
| rs780960219 | 568 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs780960219 | 568 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1196144374 | 571 | P>A | No | gnomAD | |
| rs913043415 | 571 | P>L | No |
TOPMed gnomAD |
|
| rs1948836664 | 575 | R>K | No | Ensembl | |
| rs147065047 | 577 | S>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1948836987 | 578 | S>C | No |
TOPMed gnomAD |
|
| rs1592680758 | 581 | V>G | No | Ensembl | |
| rs949864242 | 582 | A>S | No |
TOPMed gnomAD |
|
| TCGA novel | 584 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201348197 | 586 | T>A | No | Ensembl | |
| COSM938800 | 587 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1948837494 | 588 | A>V | No |
TOPMed gnomAD |
|
| rs1395755171 | 589 | S>G | No | gnomAD | |
| rs768284104 | 589 | S>N | No |
ExAC gnomAD |
|
| rs1243777008 | 590 | K>R | No | gnomAD | |
| rs1394886003 | 592 | P>L | No |
TOPMed gnomAD |
|
| rs1592680851 | 592 | P>S | No | Ensembl | |
| rs748485099 | 594 | P>L | No |
ExAC gnomAD |
|
| rs774749871 | 594 | P>T | No |
ExAC gnomAD |
|
| rs772445426 | 595 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1314647459 | 595 | T>I | No | gnomAD | |
| rs1314647459 | 595 | T>S | No | gnomAD | |
| rs1592680921 | 597 | T>P | No | Ensembl | |
| rs1231161521 | 597 | T>S | No | gnomAD | |
| rs1339365460 | 598 | P>L | No | gnomAD | |
| rs1937647328 | 598 | P>S | No | TOPMed | |
| rs905754691 | 599 | T>I | No |
TOPMed gnomAD |
|
| rs905754691 | 599 | T>K | No |
TOPMed gnomAD |
|
| TCGA novel | 599 | T>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1461399598 | 600 | I>N | No | gnomAD | |
| rs1420319439 | 600 | I>V | No | TOPMed | |
| rs761183677 | 602 | P>R | No |
ExAC gnomAD |
|
| rs944681292 | 604 | I>T | No |
TOPMed gnomAD |
|
| rs148101509 | 605 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA gnomAD |
| rs759892057 | 605 | T>I | No |
ExAC gnomAD |
|
| rs148101509 | 605 | T>S | No |
ESP ExAC gnomAD |
|
| rs765534131 | 607 | P>A | No | ExAC | |
| rs751440161 | 608 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1253954692 | 612 | V>I | No | TOPMed | |
| rs750334933 | 613 | L>* | No |
ExAC gnomAD |
|
| rs779598814 | 614 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs756076471 | 614 | D>G | No |
ExAC gnomAD |
|
| rs201431305 | 615 | T>R | No | Ensembl | |
| rs757331139 | 618 | I>V | No |
TOPMed gnomAD |
|
|
COSM3460198 rs200444584 |
619 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1318769935 | 619 | R>H | No |
TOPMed gnomAD |
|
| rs1318769935 | 619 | R>L | No |
TOPMed gnomAD |
|
| rs1277138147 | 621 | E>D | No | gnomAD | |
| rs1015652371 | 622 | P>Q | No | Ensembl | |
| rs1937653334 | 623 | M>I | No | TOPMed | |
| rs1937653182 | 623 | M>V | No |
TOPMed gnomAD |
|
| rs1937653478 | 624 | N>D | No | TOPMed | |
| rs961470910 | 625 | I>V | No | Ensembl | |
| rs201557218 | 631 | I>V | No |
ExAC TOPMed gnomAD |
|
| COSM268489 | 633 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1009886917 COSM3460200 |
633 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1937654974 | 635 | Q>* | No | Ensembl | |
| rs1937655128 | 638 | H>Q | No | Ensembl | |
| rs1937655366 | 639 | L>P | No | TOPMed | |
| rs1937655857 | 641 | K>I | No | TOPMed | |
| rs1445079135 | 642 | A>T | No | gnomAD | |
| TCGA novel | 642 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1937656676 | 643 | V>M | No | Ensembl | |
| rs998314203 | 645 | R>Q | No |
TOPMed gnomAD |
|
| rs1592681268 | 645 | R>W | No | Ensembl | |
| rs1937657521 | 646 | S>F | No | Ensembl | |
| rs199856719 | 647 | L>F | No | Ensembl | |
| rs771451999 | 648 | Q>* | No |
ExAC gnomAD |
|
| rs771451999 | 648 | Q>E | No |
ExAC gnomAD |
|
| rs1289332941 | 648 | Q>R | No | Ensembl | |
| rs769953540 | 651 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs369094443 | 651 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs769953540 | 651 | R>S | No |
ExAC TOPMed gnomAD |
|
| COSM4820082 | 653 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763175978 | 655 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs200686810 | 657 | R>P | No |
1000Genomes ExAC gnomAD |
|
| rs200686810 | 657 | R>Q | No |
1000Genomes ExAC gnomAD |
|
|
COSM252382 rs750220029 |
657 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs766277520 | 658 | E>G | No |
ExAC gnomAD |
|
| rs754508990 | 660 | A>S | No |
ExAC gnomAD |
|
| rs1348270715 | 662 | M>L | No | gnomAD | |
| rs200110308 | 662 | M>R | No |
1000Genomes ExAC |
|
| rs1348270715 | 662 | M>V | No | gnomAD | |
| rs778054016 | 663 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1937661831 | 665 | K>E | No |
TOPMed gnomAD |
|
| rs747373437 | 667 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs375672599 | 669 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs375672599 | 669 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1937662618 | 671 | M>T | No | Ensembl | |
| TCGA novel | 673 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1197214226 | 674 | I>F | No | gnomAD | |
| TCGA novel | 676 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1266809308 | 679 | S>A | No |
TOPMed gnomAD |
|
| COSM3460201 | 679 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 683 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs957088230 | 688 | I>T | No |
TOPMed gnomAD |
|
|
COSM1361246 rs1363511738 |
689 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs746325904 | 693 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs763144810 | 700 | Q>* | No | ExAC | |
| rs751131276 | 704 | V>A | No |
ExAC gnomAD |
|
| rs201936676 | 705 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs868324185 | 707 | A>S | No | Ensembl | |
| TCGA novel | 710 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs139319862 | 713 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs79335280 | 713 | Y>S | No | Ensembl | |
| TCGA novel | 718 | A>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs904439163 | 719 | M>V | No |
TOPMed gnomAD |
|
| TCGA novel | 721 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs112529809 | 725 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs112529809 | 725 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs1297601528 | 726 | K>N | No | gnomAD | |
| TCGA novel | 728 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1938024912 | 729 | N>Y | No | Ensembl | |
| rs774660181 | 730 | E>* | No |
ExAC gnomAD |
|
| TCGA novel | 732 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1938025457 | 738 | A>S | No | Ensembl | |
| rs1938025796 | 739 | A>S | No | TOPMed | |
| rs1938025984 | 740 | T>A | No | Ensembl | |
| rs1938026145 | 740 | T>N | No | Ensembl | |
| rs748263015 | 741 | F>S | No |
ExAC gnomAD |
|
|
rs1160828139 COSM3460202 |
743 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs200219306 | 746 | A>G | No | Ensembl | |
| rs1324660893 | 747 | M>T | No | gnomAD | |
| rs199504080 | 753 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1938179636 | 753 | D>H | No | TOPMed | |
| COSM4924307 | 755 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 755 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1486252831 | 756 | V>L | No | gnomAD | |
| rs1364373686 | 760 | D>E | No |
TOPMed gnomAD |
|
| rs1938180368 | 760 | D>H | No |
TOPMed gnomAD |
|
| rs751235193 | 761 | E>D | No |
ExAC gnomAD |
|
| rs1938180856 | 762 | M>I | No | Ensembl | |
| rs1336962124 | 763 | Q>R | No | gnomAD | |
| rs375067221 | 767 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs768053496 | 768 | A>D | No |
ExAC gnomAD |
|
| rs768053496 | 768 | A>G | No |
ExAC gnomAD |
|
| rs2136280432 | 770 | E>D | No | Ensembl | |
| COSM5987300 | 770 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1290366943 | 771 | D>N | No | gnomAD | |
| rs750796204 | 773 | K>R | No |
ExAC gnomAD |
|
| COSM1361248 | 774 | K>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1028462141 | 774 | K>T | No | TOPMed | |
|
VAR_069060 rs200845476 |
778 | T>A | No |
UniProt TOPMed dbSNP gnomAD |
|
| COSM468280 | 779 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM32836 rs749713234 |
781 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs201842580 | 781 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs201842580 | 781 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1208308896 | 782 | M>K | No | gnomAD | |
| rs1170596357 | 784 | I>M | No |
TOPMed gnomAD |
|
| rs1260328579 | 789 | A>D | No | gnomAD | |
|
rs200892811 COSM4041424 |
789 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs199987993 | 795 | E>G | No |
ExAC gnomAD |
|
| TCGA novel | 795 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1592693698 | 796 | D>G | No | Ensembl | |
| TCGA novel | 796 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1592693711 | 799 | F>V | No | Ensembl | |
| rs1160849845 | 800 | D>N | No | gnomAD | |
| rs201080623 | 801 | H>Q | No | Ensembl | |
| rs1429840526 | 801 | H>R | No |
TOPMed gnomAD |
|
| rs745628029 | 801 | H>Y | No |
ExAC gnomAD |
|
| rs1287045247 | 803 | Q>* | No | gnomAD | |
| rs769432193 | 804 | S>P | No |
ExAC gnomAD |
|
| rs1448743194 | 805 | R>* | No | Ensembl | |
| rs1448743194 | 805 | R>G | No | Ensembl | |
| rs367674204 | 805 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs202245331 | 806 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs202245331 | 806 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs773727829 | 806 | R>H | No |
ExAC gnomAD |
|
| rs146453819 | 807 | S>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1309413443 | 807 | S>N | No | TOPMed | |
| rs1938189241 | 808 | K>N | No | TOPMed | |
| rs767249691 | 809 | G>D | No | ExAC | |
| rs1286625118 | 809 | G>S | No | gnomAD | |
| rs761147593 | 812 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs1938190226 | 814 | S>N | No | TOPMed | |
| rs1284461433 | 816 | I>L | No | gnomAD | |
| rs200867915 | 816 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs201830478 | 817 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201830478 | 817 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs187011016 | 818 | S>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs752748237 | 818 | S>R | No |
ExAC gnomAD |
|
| rs758654796 | 820 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1399864305 | 821 | V>G | No | gnomAD | |
| rs1200591989 | 823 | G>E | No |
TOPMed gnomAD |
|
| rs1397606289 | 825 | A>S | No |
TOPMed gnomAD |
|
| rs1425176600 | 826 | S>* | No |
TOPMed gnomAD |
|
| rs1592693953 | 828 | T>A | No | TOPMed | |
| rs1592693953 | 828 | T>P | No | TOPMed | |
| rs1938193844 | 829 | V>G | No | TOPMed | |
| rs142080852 | 829 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1281482219 | 830 | P>S | No | gnomAD | |
| rs1281482219 | 830 | P>T | No | gnomAD | |
| rs1938194364 | 833 | D>E | No | TOPMed | |
| rs899906832 | 833 | D>N | No | Ensembl | |
| rs769644251 | 834 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1938194830 | 835 | Y>F | No | TOPMed | |
| rs151138129 | 836 | L>F | No |
ESP ExAC gnomAD |
|
| rs151138129 | 836 | L>V | No |
ESP ExAC gnomAD |
|
| rs1011645391 | 837 | L>P | No | gnomAD | |
| rs1011645391 | 837 | L>R | No | gnomAD | |
| rs201245166 | 838 | H>R | No | TOPMed | |
| rs1223805173 | 839 | S>N | No | gnomAD | |
| rs1291765244 | 839 | S>R | No | gnomAD | |
| rs1167377451 | 840 | Q>H | No | TOPMed | |
| rs1449375322 | 840 | Q>K | No | gnomAD | |
| rs1206060064 | 842 | P>S | No | gnomAD | |
| COSM4936500 | 843 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1173835809 | 845 | P>A | No | gnomAD | |
| rs201938950 | 846 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1592694099 | 847 | I>V | No | Ensembl | |
| rs200371393 | 848 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs761440526 COSM1162433 |
848 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs760341874 | 849 | V>G | No |
ExAC gnomAD |
|
|
COSM468281 rs1555170325 |
852 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs140068155 | 854 | Q>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1198217917 | 854 | Q>R | No | TOPMed | |
| rs1162833210 | 857 | R>K | No | gnomAD | |
| rs1938236514 | 858 | Q>* | No | gnomAD | |
| TCGA novel | 858 | Q>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1448539441 | 861 | P>R | No | gnomAD | |
| rs1938236972 | 862 | S>Y | No | TOPMed | |
| rs1273957172 | 863 | L>F | No | TOPMed | |
| COSM3460203 | 865 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1938237541 | 866 | Q>E | No | Ensembl | |
| rs751625209 | 867 | S>N | No |
ExAC gnomAD |
|
|
COSM6072456 rs764225109 |
867 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC gnomAD NCI-TCGA Cosmic |
| rs1431570417 | 868 | R>C | No | gnomAD | |
| rs111567892 | 868 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM5721914 | 869 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1172217593 | 870 | R>K | No | gnomAD | |
| TCGA novel | 870 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1938238791 | 873 | G>E | No | TOPMed | |
| rs1399364798 | 873 | G>R | No | TOPMed | |
| rs201853036 | 874 | A>S | No |
TOPMed gnomAD |
|
| rs201853036 | 874 | A>T | No |
TOPMed gnomAD |
|
| rs1938239115 | 874 | A>V | No | Ensembl | |
| rs1938239360 | 875 | S>Y | No | gnomAD | |
| rs778786985 | 876 | Y>D | No |
ExAC gnomAD |
|
| rs758400791 | 880 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs147946184 | 881 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 882 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1205545202 | 883 | V>A | No | gnomAD | |
| rs1938240864 | 884 | P>A | No | gnomAD | |
| COSM5527359 | 884 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1592695393 | 885 | P>L | No | Ensembl | |
| rs1938241450 | 887 | P>S | No | TOPMed | |
| rs994501899 | 888 | T>I | No | gnomAD | |
| rs1026422872 | 889 | S>A | No | Ensembl | |
| rs1938242083 | 890 | T>A | No | TOPMed | |
| rs1938242083 | 890 | T>S | No | TOPMed | |
| COSM3460204 | 892 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1938242760 | 897 | G>D | No | Ensembl | |
| TCGA novel | 898 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770147066 | 898 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs775825494 | 899 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1005858759 | 899 | P>L | No |
TOPMed gnomAD |
|
| rs775825494 | 899 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs775825494 | 899 | P>T | No |
ExAC TOPMed gnomAD |
|
| COSM4041425 | 900 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1016876023 | 900 | S>F | No | TOPMed | |
| rs1938243768 | 905 | I>T | No | TOPMed | |
| rs1938244066 | 906 | Q>L | No | Ensembl | |
| rs950480628 | 907 | G>A | No |
TOPMed gnomAD |
|
| COSM3398672 | 907 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1298704531 | 908 | H>D | No | gnomAD | |
| rs1337213894 | 908 | H>R | No |
TOPMed gnomAD |
|
| rs1298704531 | 908 | H>Y | No | gnomAD | |
| rs774882871 | 909 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs764562180 | 909 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1219549059 | 910 | L>F | No |
TOPMed gnomAD |
|
| rs1276120971 | 911 | S>N | No |
TOPMed gnomAD |
|
| rs1276120971 | 911 | S>T | No |
TOPMed gnomAD |
|
| rs767577430 | 912 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1217882321 | 913 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1379843185 | 913 | E>A | No |
TOPMed gnomAD |
|
| rs532212955 | 915 | R>G | No |
1000Genomes ExAC gnomAD |
|
| rs1421053536 | 915 | R>K | No | gnomAD | |
| COSM4041426 | 917 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752521721 | 917 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs752521721 | 917 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs141754177 | 918 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM3460205 | 920 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756728997 | 922 | D>N | No |
ExAC gnomAD |
|
| rs766680668 | 923 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs766680668 | 923 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs200887827 | 925 | Q>* | No |
ExAC gnomAD |
|
| rs374128485 | 928 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1939576096 | 928 | A>V | No | TOPMed | |
| rs1309332367 | 929 | S>F | No |
TOPMed gnomAD |
|
| rs919170528 | 930 | V>E | No |
TOPMed gnomAD |
|
| rs200305362 | 930 | V>I | No |
TOPMed gnomAD |
|
|
rs757213231 COSM1184702 |
931 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs200338613 | 932 | P>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1939577232 | 933 | Q>* | No | TOPMed | |
| rs1939577232 | 933 | Q>E | No | TOPMed | |
| rs755803723 | 934 | C>R | No |
ExAC gnomAD |
|
| rs1939577705 | 936 | Q>P | No | TOPMed | |
| rs202093248 | 938 | A>T | No |
ExAC TOPMed gnomAD |
|
| COSM3460206 | 938 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1705361 | 939 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs769016012 | 939 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1038052156 | 940 | R>K | No | gnomAD | |
| rs898210192 | 941 | Q>K | No |
TOPMed gnomAD |
|
| rs1176710096 | 941 | Q>R | No | gnomAD | |
| rs1939579750 | 942 | D>H | No | Ensembl | |
| rs1939579950 | 944 | P>S | No | Ensembl | |
| rs2136331810 | 947 | S>G | No | Ensembl | |
| rs1160962924 | 948 | P>A | No | gnomAD | |
| rs1160962924 | 948 | P>T | No | gnomAD | |
| rs1296281985 | 949 | D>E | No | gnomAD | |
| rs753556215 | 949 | D>G | No |
ExAC gnomAD |
|
| rs754648158 | 950 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs754648158 | 950 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1940021598 | 951 | A>S | No | TOPMed | |
| rs749237150 | 951 | A>V | No |
TOPMed gnomAD |
|
| rs1275627940 | 952 | L>F | No | gnomAD | |
| rs1940021959 | 953 | P>T | No | Ensembl | |
| rs779197304 | 955 | E>D | No |
ExAC gnomAD |
|
| rs1940022282 | 955 | E>K | No |
TOPMed gnomAD |
|
| rs1565706697 | 956 | Q>P | No | TOPMed | |
| rs1565706697 | 956 | Q>R | No | TOPMed | |
| rs201836822 | 957 | P>T | No |
ESP TOPMed gnomAD |
|
| rs748517592 | 958 | H>L | No |
ExAC gnomAD |
|
| rs748517592 | 958 | H>R | No |
ExAC gnomAD |
|
| rs1427799637 | 959 | S>C | No | Ensembl | |
| rs1427799637 | 959 | S>F | No | Ensembl | |
| rs138241223 | 960 | S>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs778361558 | 963 | C>G | No |
ExAC TOPMed gnomAD |
|
| rs778361558 | 963 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs746149233 | 964 | A>D | No |
ExAC gnomAD |
|
|
rs200603103 COSM166457 |
964 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs770033744 | 965 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs770033744 | 965 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs201572851 | 968 | C>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs201572851 | 968 | C>Y | No |
1000Genomes ExAC gnomAD |
|
| rs2136348952 | 969 | L>P | No | Ensembl | |
| TCGA novel | 972 | P>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1940024768 | 972 | P>R | No | Ensembl | |
| rs1565706798 | 972 | P>S | No | Ensembl | |
| rs767158928 | 973 | P>R | No |
ExAC gnomAD |
|
| rs1438920371 | 973 | P>S | No | gnomAD | |
| rs1592737534 | 974 | H>P | No | Ensembl | |
| rs1387053896 | 975 | P>S | No | gnomAD | |
| rs1940026147 | 976 | P>L | No | Ensembl | |
| rs1940025986 | 976 | P>Q | No |
TOPMed gnomAD |
|
| rs1487455730 | 976 | P>S | No | TOPMed | |
| rs750962254 | 976 | P>S | No | gnomAD |
No associated diseases with Q96G01
1 regional properties for Q96G01
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 16 - 169 | IPR005225 |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| secretory vesicle | A cytoplasmic, membrane bound vesicle that is capable of fusing to the plasma membrane to release its contents into the extracellular space. |
| trans-Golgi network | The network of interconnected tubular and cisternal structures located within the Golgi apparatus on the side distal to the endoplasmic reticulum, from which secretory vesicles emerge. The trans-Golgi network is important in the later stages of protein secretion where it is thought to play a key role in the sorting and targeting of secreted proteins to the correct destination. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| cytoskeletal anchor activity | The binding activity of a protein that brings together a cytoskeletal protein (either a microtubule or actin filament, spindle pole body, or protein directly bound to them) and one or more other molecules, permitting them to function in a coordinated way. |
| dynactin binding | Binding to a dynactin complex; a large protein complex that activates dynein-based motor activity. |
| dynein complex binding | Binding to a dynein complex, a protein complex that contains two or three dynein heavy chains and several light chains, and has microtubule motor activity. |
| dynein intermediate chain binding | Binding to an intermediate chain of the dynein complex. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| proteinase activated receptor binding | Binding to a proteinase activated receptor. |
| small GTPase binding | Binding to a small monomeric GTPase. |
| structural constituent of cytoskeleton | The action of a molecule that contributes to the structural integrity of a cytoskeletal structure. |
14 GO annotations of biological process
| Name | Definition |
|---|---|
| anatomical structure morphogenesis | The process in which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form. |
| intracellular mRNA localization | Any process in which mRNA is transported to, or maintained in, a specific location within the cell. |
| microtubule anchoring at microtubule organizing center | Any process in which a microtubule is maintained in a specific location in a cell by attachment to a microtubule organizing center. |
| minus-end-directed organelle transport along microtubule | The directed movement of an organelle towards the minus end of a microtubule, mediated by motor proteins. This process begins with the attachment of an organelle to a microtubule, and ends when the organelle reaches its final destination. |
| negative regulation of phospholipase C activity | Any process that stops, prevents or reduces the frequency, rate or extent of phospholipase C activity. |
| negative regulation of phospholipase C-activating G protein-coupled receptor signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of phospholipase C-activating G protein-coupled receptor signaling pathway. |
| positive regulation of protein localization to centrosome | Any process that activates or increases the frequency, rate or extent of protein localization to centrosome. |
| positive regulation of receptor-mediated endocytosis | Any process that activates or increases the frequency, rate or extent of receptor mediated endocytosis, the uptake of external materials by cells, utilizing receptors to ensure specificity of transport. |
| protein localization to organelle | A process in which a protein is transported to, or maintained in, a location within an organelle. |
| regulation of microtubule cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising microtubules and their associated proteins. |
| regulation of proteinase activated receptor activity | Any process that modulates the frequency, rate or extent of proteinase activated receptor activity. |
| RNA processing | Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules. |
| stress granule assembly | The aggregation, arrangement and bonding together of proteins and RNA molecules to form a stress granule. |
| viral process | A multi-organism process in which a virus is a participant. The other participant is the host. Includes infection of a host cell, replication of the viral genome, and assembly of progeny virus particles. In some cases the viral genetic material may integrate into the host genome and only subsequently, under particular circumstances, 'complete' its life cycle. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P16568 | BicD | Protein bicaudal D | Drosophila melanogaster (Fruit fly) | EV |
| Q8TD16 | BICD2 | Protein bicaudal D homolog 2 | Homo sapiens (Human) | SS |
| Q921C5 | Bicd2 | Protein bicaudal D homolog 2 | Mus musculus (Mouse) | SS |
| Q8BR07 | Bicd1 | Protein bicaudal D homolog 1 | Mus musculus (Mouse) | EV |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAEEVLQTV | DHYKTEIERL | TKELTETTHE | KIQAAEYGLV | VLEEKLTLKQ | QYDELEAEYD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SLKQELEQLK | EAFGQSFSIH | RKVAEDGETR | EETLLQESAS | KEAYYLGKIL | EMQNELKQSR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AVVTNVQAEN | ERLTAVVQDL | KENNEMVELQ | RIRMKDEIRE | YKFREARLLQ | DYTELEEENI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TLQKLVSTLK | QNQVEYEGLK | HEIKRFEEET | VLLNSQLEDA | IRLKEIAEHQ | LEEALETLKN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EREQKNNLRK | ELSQYISLND | NHISISVDGL | KFAEDGSEPN | NDDKMNGHIH | GPLVKLNGDY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RTPTLRKGES | LNPVSDLFSE | LNISEIQKLK | QQLMQVEREK | AILLANLQES | QTQLEHTKGA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LTEQHERVHR | LTEHVNAMRG | LQSSKELKAE | LDGEKGRDSG | EEAHDYEVDI | NGLEILECKY |
| 430 | 440 | 450 | 460 | 470 | 480 |
| RVAVTEVIDL | KAEIKALKEK | YNKSVENYTD | EKAKYESKIQ | MYDEQVTSLE | KTTKESGEKM |
| 490 | 500 | 510 | 520 | 530 | 540 |
| AHMEKELQKM | TSIANENHST | LNTAQDELVT | FSEELAQLYH | HVCLCNNETP | NRVMLDYYRQ |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SRVTRSGSLK | GPDDPRGLLS | PRLARRGVSS | PVETRTSSEP | VAKESTEASK | EPSPTKTPTI |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SPVITAPPSS | PVLDTSDIRK | EPMNIYNLNA | IIRDQIKHLQ | KAVDRSLQLS | RQRAAARELA |
| 670 | 680 | 690 | 700 | 710 | 720 |
| PMIDKDKEAL | MEEILKLKSL | LSTKREQIAT | LRAVLKANKQ | TAEVALANLK | NKYENEKAMV |
| 730 | 740 | 750 | 760 | 770 | 780 |
| TETMTKLRNE | LKALKEDAAT | FSSLRAMFAT | RCDEYVTQLD | EMQRQLAAAE | DEKKTLNTLL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| RMAIQQKLAL | TQRLEDLEFD | HEQSRRSKGK | LGKSKIGSPK | VSGEASVTVP | TIDTYLLHSQ |
| 850 | 860 | 870 | 880 | 890 | 900 |
| GPQTPNIRVS | SGTQRKRQFS | PSLCDQSRPR | TSGASYLQNL | LRVPPDPTST | ESFLLKGPPS |
| 910 | 920 | 930 | 940 | 950 | 960 |
| MSEFIQGHRL | SKEKRLTVAP | PDCQQPAASV | PPQCSQLAGR | QDCPTVSPDT | ALPEEQPHSS |
| 970 | |||||
| SQCAPLHCLS | KPPHP |