Q96EB6
Gene name |
SIRT1 |
Protein name |
NAD-dependent protein deacetylase sirtuin-1 |
Names |
hSIRT1, NAD-dependent protein deacylase sirtuin-1, Regulatory protein SIR2 homolog 1, SIR2-like protein 1, hSIR2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:23411 |
EC number |
2.3.1.286: Transferring groups other than amino-acyl groups |
Protein Class |
|
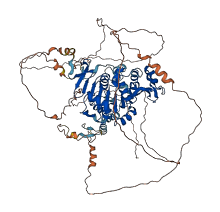
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

10 structures for Q96EB6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4I5I | X-ray | 250 A | A/B | 241-516 | PDB |
| 4IF6 | X-ray | 225 A | PDB | ||
| 4IG9 | X-ray | 264 A | PDB | ||
| 4KXQ | X-ray | 185 A | PDB | ||
| 4ZZH | X-ray | 310 A | A | 183-505 | PDB |
| 4ZZI | X-ray | 273 A | A | 183-505 | PDB |
| 4ZZJ | X-ray | 274 A | A | 183-505 | PDB |
| 5BTR | X-ray | 320 A | A/B/C | 143-665 | PDB |
| 8ANB | X-ray | 164 A | P | 665-675 | PDB |
| AF-Q96EB6-F1 | Predicted | AlphaFoldDB |
562 variants for Q96EB6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs587776957 RCV000041965 CA143680 |
107 | L>P | Variant of unknown significance [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1165843217 CA377035040 |
2 | A>V | No |
ClinGen gnomAD |
|
|
CA208364030 rs35671182 VAR_025148 CA377035047 |
3 | D>E | No |
ClinGen TOPMed UniProt dbSNP |
|
|
CA377035051 rs1589064382 |
4 | E>A | No |
ClinGen Ensembl |
|
|
rs955794368 CA208364034 |
5 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA377035069 rs1485737596 |
7 | L>F | No |
ClinGen TOPMed |
|
|
CA377035081 rs1291063294 |
9 | L>F | No |
ClinGen TOPMed |
|
|
rs966205195 CA208364062 |
11 | P>A | No |
ClinGen Ensembl |
|
|
CA208364084 rs200005116 |
13 | G>C | No |
ClinGen TOPMed |
|
|
CA5520972 rs201230502 |
14 | S>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377035119 rs1368006921 |
15 | P>L | No |
ClinGen TOPMed |
|
|
CA377035114 rs1381917502 |
15 | P>T | No |
ClinGen TOPMed |
|
|
CA208364101 rs995901311 |
16 | S>L | No |
ClinGen Ensembl |
|
|
CA377035122 rs1422614899 |
16 | S>T | No |
ClinGen TOPMed |
|
|
CA377035134 rs1431102999 |
18 | A>V | No |
ClinGen TOPMed |
|
|
CA377035164 rs1323967226 |
23 | E>K | No |
ClinGen gnomAD |
|
|
rs1268445241 CA377035176 |
24 | A>G | No |
ClinGen TOPMed |
|
|
rs1489985293 CA377035189 |
26 | S>L | No |
ClinGen TOPMed |
|
|
rs1202064844 CA377035184 |
26 | S>T | No |
ClinGen TOPMed |
|
|
CA377035198 rs1589064466 |
27 | S>F | No |
ClinGen Ensembl |
|
|
CA208364111 rs899362289 |
27 | S>P | No |
ClinGen Ensembl |
|
|
rs920003355 CA208364124 |
28 | P>R | No |
ClinGen TOPMed |
|
|
CA208364125 rs948757989 |
30 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA377035262 rs1589064475 |
32 | P>R | No |
ClinGen Ensembl |
|
|
rs1344714636 CA377035326 |
37 | P>A | No |
ClinGen TOPMed |
|
|
CA5520974 rs548590752 |
37 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA377035387 rs1365874384 |
41 | G>A | No |
ClinGen gnomAD |
|
|
CA377035395 rs1589064501 |
42 | P>S | No |
ClinGen Ensembl |
|
|
CA208364133 rs1053224730 |
44 | L>H | No |
ClinGen TOPMed gnomAD |
|
|
CA377035439 rs1466187737 |
45 | E>D | No |
ClinGen TOPMed |
|
|
CA377035427 rs1166840400 |
45 | E>Q | No |
ClinGen TOPMed |
|
|
rs1379015674 CA377035444 |
46 | R>Q | No |
ClinGen TOPMed |
|
|
rs568432780 CA208364141 |
48 | P>L | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs1011345346 CA208364137 |
48 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA377035482 rs1183497853 |
49 | G>D | No |
ClinGen TOPMed |
|
|
rs1440092527 CA377035568 |
56 | P>R | No |
ClinGen TOPMed |
|
|
CA377035575 rs1195315722 |
57 | E>A | No |
ClinGen TOPMed |
|
|
rs1021816786 CA208364144 |
57 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1483663888 CA377035588 |
58 | R>C | No |
ClinGen TOPMed |
|
|
rs1227716359 CA377035612 |
59 | E>D | No |
ClinGen gnomAD |
|
|
rs1265229401 CA377035609 |
59 | E>V | No |
ClinGen TOPMed |
|
|
CA377035636 rs1446726608 |
61 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1446726608 CA377035631 |
61 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA208364157 rs964485281 |
63 | A>V | No |
ClinGen TOPMed |
|
|
rs1333930827 CA377035668 |
64 | A>T | No |
ClinGen TOPMed |
|
|
rs531048058 CA5520976 |
66 | G>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs531048058 CA377035699 |
66 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1450691401 CA377035737 |
68 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1450691401 CA377035739 |
68 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
CA377035742 rs1159536580 |
69 | G>S | No |
ClinGen TOPMed |
|
|
rs751564985 CA5520977 |
70 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs907725955 CA208364197 |
71 | A>V | No |
ClinGen TOPMed |
|
|
CA377035790 rs1461061909 |
72 | A>V | No |
ClinGen TOPMed |
|
|
CA377035855 rs1206687979 |
77 | R>L | No |
ClinGen TOPMed |
|
|
CA377035848 rs1459511345 |
77 | R>W | No |
ClinGen gnomAD |
|
|
rs864309601 RCV000202791 CA248985 |
78 | E>G | No |
ClinGen ClinVar dbSNP gnomAD |
|
|
rs1278877193 CA377035860 |
78 | E>K | No |
ClinGen TOPMed |
|
|
CA377035883 rs1564880865 |
79 | A>G | No |
ClinGen Ensembl |
|
|
CA377035898 rs1451404296 |
80 | E>D | No |
ClinGen TOPMed |
|
|
rs1388942211 CA377035907 |
81 | A>S | No |
ClinGen TOPMed |
|
|
rs1383333215 CA377035935 |
83 | A>G | No |
ClinGen TOPMed |
|
|
rs920039980 CA208364211 |
83 | A>T | No |
ClinGen TOPMed |
|
|
rs1425887328 CA377035951 |
84 | A>V | No |
ClinGen TOPMed |
|
|
rs1384549244 CA377035966 |
86 | A>T | No |
ClinGen gnomAD |
|
|
rs1181845230 CA377036031 |
90 | Q>P | No |
ClinGen TOPMed |
|
|
CA377036049 rs1378803687 |
91 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1455404263 CA377036038 |
91 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA377036055 rs1478153249 |
92 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1478153249 CA377036057 |
92 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA208364221 rs948830328 |
92 | A>V | No |
ClinGen TOPMed |
|
|
rs1283355068 CA377036070 |
93 | Q>H | No |
ClinGen TOPMed |
|
|
rs1171595277 CA377036074 |
94 | A>S | No |
ClinGen gnomAD |
|
|
CA208364229 rs1044796885 |
95 | T>I | No |
ClinGen TOPMed |
|
|
rs767360333 CA5520979 |
95 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs550317521 CA5520980 |
97 | A>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs990065260 CA208364234 |
100 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs199624804 CA208364230 |
100 | E>G | No |
ClinGen Ensembl |
|
|
CA208364251 rs892030271 |
101 | G>E | No |
ClinGen TOPMed |
|
|
rs913135320 CA208364245 |
101 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA377036166 rs1390008266 |
102 | D>H | No |
ClinGen gnomAD |
|
|
rs978078516 CA208364257 |
103 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA377036216 rs1406033790 |
105 | P>L | No |
ClinGen TOPMed |
|
|
CA208364261 rs927927572 |
106 | G>V | No |
ClinGen Ensembl |
|
|
CA377036257 rs1437499917 |
108 | Q>H | No |
ClinGen gnomAD |
|
|
rs1278542503 CA377036267 |
109 | G>D | No |
ClinGen gnomAD |
|
|
rs1589064688 CA377036261 |
109 | G>S | No |
ClinGen Ensembl |
|
|
rs868722110 CA208364268 |
110 | P>A | No |
ClinGen Ensembl |
|
|
rs1326686646 CA377036281 |
110 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1326686646 CA377036279 |
110 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1282830986 CA377036294 |
111 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1321218486 CA377036297 |
112 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1321218486 CA377036299 |
112 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
rs1200315460 CA377036321 |
113 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1465739818 CA377036308 |
113 | E>K | No |
ClinGen gnomAD |
|
|
CA5520982 rs182199697 COSM3686825 |
114 | P>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs1254698465 CA377036328 |
114 | P>S | No |
ClinGen gnomAD |
|
|
CA208364270 rs900251478 |
115 | P>A | No |
ClinGen TOPMed |
|
|
rs930809097 CA208364275 |
115 | P>L | No |
ClinGen TOPMed |
|
|
CA208364281 rs886702977 |
116 | L>M | No |
ClinGen TOPMed gnomAD |
|
|
CA5520984 rs756786838 |
117 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA208364286 rs865785083 |
119 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA377036393 rs865785083 |
119 | N>T | No |
ClinGen TOPMed gnomAD |
|
|
rs779354477 CA5520985 |
120 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA208364289 rs1030015286 |
120 | L>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1479697482 CA377036447 |
122 | D>E | No |
ClinGen gnomAD |
|
|
CA377036433 rs1266282487 |
122 | D>N | No |
ClinGen gnomAD |
|
|
CA377036488 rs1184771827 |
124 | D>E | No |
ClinGen gnomAD |
|
|
CA377036479 rs1412525648 |
124 | D>N | No |
ClinGen gnomAD |
|
|
CA377036526 rs1413092045 |
126 | D>G | No |
ClinGen gnomAD |
|
|
rs1453683523 CA377036530 |
127 | D>N | No |
ClinGen gnomAD |
|
|
CA208364304 rs865826245 |
128 | E>K | No |
ClinGen Ensembl |
|
|
rs746381892 CA5520989 |
129 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA377036596 rs1406007212 |
131 | E>G | No |
ClinGen TOPMed gnomAD |
|
|
rs868088559 CA208364310 |
131 | E>K | No |
ClinGen Ensembl |
|
|
CA377036612 rs1427006382 |
132 | E>K | No |
ClinGen TOPMed |
|
|
CA377036625 rs1198565310 |
133 | E>K | No |
ClinGen TOPMed |
|
|
CA5520993 rs775978922 |
136 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA208364339 rs867689409 |
136 | A>T | No |
ClinGen Ensembl |
|
|
rs775978922 CA377036683 |
136 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377036714 rs1396067593 |
139 | A>E | No |
ClinGen gnomAD |
|
|
rs1023430455 CA208364351 |
139 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
rs142378619 CA208366179 |
144 | D>G | No |
ClinGen ESP TOPMed |
|
|
rs1024951677 CA208366182 |
145 | N>H | No |
ClinGen TOPMed |
|
|
rs772106776 CA5521016 |
146 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs748384143 CA5521017 |
147 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377038529 rs773494159 |
148 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA208366210 rs978488168 |
149 | G>D | No |
ClinGen TOPMed |
|
| TCGA novel | 149 | G>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs776925394 CA5521020 |
150 | D>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 151 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377038608 rs1451869566 |
154 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
CA377038614 rs1173732078 |
154 | T>S | No |
ClinGen gnomAD |
|
|
CA377038620 rs1441322674 |
155 | N>D | No |
ClinGen TOPMed |
|
|
CA5521021 rs767540399 |
158 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs200660028 CA5521022 |
159 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA5521023 rs760482544 |
159 | S>Y | No |
ClinGen ExAC |
|
|
rs763883280 CA5521024 |
160 | C>S | No |
ClinGen ExAC gnomAD |
|
|
rs753433479 CA5521025 |
160 | C>W | No |
ClinGen ExAC gnomAD |
|
|
rs1466540364 CA377038697 |
160 | C>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs1323755686 CA377038724 |
162 | S>G | No |
ClinGen TOPMed |
|
|
rs1460870553 CA377038735 |
162 | S>R | No |
ClinGen TOPMed |
|
|
CA5521026 rs761508157 |
163 | D>E | No |
ClinGen ExAC |
|
|
rs1305391134 CA377038787 |
166 | D>N | No |
ClinGen gnomAD |
|
|
rs144124002 RCV000938820 CA5521028 |
170 | H>R | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA208366276 rs989670245 |
171 | A>T | No |
ClinGen TOPMed |
|
|
rs758805613 CA5521029 |
174 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA5521030 rs565217830 |
177 | T>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs752035789 CA5521031 |
178 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA5521033 rs200805107 |
179 | R>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA377038993 rs1426968560 |
181 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA5521035 rs547102478 |
181 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1426968560 CA377038995 |
181 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA377039019 rs1174240434 |
182 | I>M | No |
ClinGen gnomAD |
|
|
rs749341245 CA5521037 |
182 | I>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 183 | G>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377039072 rs1223860015 |
183 | G>D | No |
ClinGen gnomAD |
|
|
rs201841214 CA208367223 |
186 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1253732619 CA377039113 |
189 | Q>R | No |
ClinGen gnomAD |
|
|
CA5521067 rs772852024 |
192 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1391705142 CA377039142 |
193 | M>I | No |
ClinGen Ensembl |
|
|
CA5521069 rs200994303 |
193 | M>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs762526864 CA5521068 |
193 | M>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 194 | I>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1292146988 CA377039145 |
194 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA377039162 rs1364332887 |
196 | T>I | No |
ClinGen gnomAD |
|
|
CA377039178 rs773774776 |
199 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA377039202 rs1352430885 |
203 | K>E | No |
ClinGen TOPMed |
|
|
rs375988661 CA208367300 COSM3723550 |
207 | P>L | upper_aerodigestive_tract [Cosmic] | No |
ClinGen cosmic curated ESP TOPMed gnomAD |
|
CA377039245 rs1387726935 |
209 | T>A | No |
ClinGen gnomAD |
|
|
rs756483371 CA208367309 |
209 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA5521074 rs756483371 |
209 | T>R | No |
ClinGen ExAC gnomAD |
|
|
rs201862792 CA377039253 |
210 | I>M | No |
ClinGen TOPMed |
|
|
rs369379188 CA5521076 |
210 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA208367329 rs201668639 |
211 | P>L | No |
ClinGen Ensembl |
|
|
CA5521077 rs201007799 |
211 | P>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 214 | E>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377039295 rs1438249152 |
217 | D>N | No |
ClinGen gnomAD |
|
|
rs201583175 CA208367341 |
218 | M>T | No |
ClinGen Ensembl |
|
|
rs745979871 CA5521079 |
218 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs199716245 CA208367344 |
219 | T>R | No |
ClinGen TOPMed gnomAD |
|
|
CA377039327 rs1482498508 |
222 | Q>K | No |
ClinGen TOPMed |
|
|
rs1442219001 CA377039336 |
223 | I>L | No |
ClinGen gnomAD |
|
|
rs769519031 CA5521083 |
223 | I>S | No |
ClinGen ExAC gnomAD |
|
|
CA377039350 rs1328620249 |
225 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA5521084 rs559057403 |
225 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1353054670 CA377039355 |
226 | N>D | No |
ClinGen gnomAD |
|
|
rs1589068201 CA377039365 |
227 | I>T | No |
ClinGen Ensembl |
|
|
rs1382176398 CA377039396 |
232 | P>Q | No |
ClinGen gnomAD |
|
|
rs750787952 CA208367394 |
232 | P>S | No |
ClinGen gnomAD |
|
|
rs750787952 CA377039394 |
232 | P>T | No |
ClinGen gnomAD |
|
|
CA377039403 rs1450537164 |
233 | K>R | No |
ClinGen gnomAD |
|
|
CA208367402 rs879180328 |
234 | R>K | No |
ClinGen gnomAD |
|
|
CA377039420 rs1270657663 |
235 | K>N | No |
ClinGen TOPMed gnomAD |
|
| rs1564883215 | 237 | R>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs879037598 CA208367418 |
237 | R>K | No |
ClinGen gnomAD |
|
|
CA5521088 rs199618656 |
240 | I>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5521087 rs199618656 |
240 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5521090 rs776084517 |
243 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA5521089 rs200711525 |
243 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA5521092 rs201479376 |
245 | D>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs761327480 CA5521091 |
245 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs1471141881 CA377039562 |
256 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA377039571 rs1405977818 |
257 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA5521095 rs765326045 |
257 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs750671807 CA5521096 |
259 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA377039586 rs1394031280 |
260 | T>A | No |
ClinGen TOPMed |
|
| TCGA novel | 264 | V>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1564884263 CA377039774 |
266 | V>L | No |
ClinGen Ensembl |
|
|
rs865778460 CA208368698 |
267 | S>L | No |
ClinGen Ensembl |
|
| TCGA novel | 270 | I>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377039826 rs1479647397 |
270 | I>V | No |
ClinGen gnomAD |
|
| TCGA novel | 273 | F>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs779685735 CA5521127 |
275 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs776933716 CA5521130 |
277 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA377039967 rs1589070016 |
279 | I>V | No |
ClinGen Ensembl |
|
|
rs1367456169 CA377040013 |
282 | R>C | No |
ClinGen gnomAD |
|
|
rs762393274 CA5521132 COSM919611 |
282 | R>H | Variant assessed as Somatic; 9.24e-05 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs762393274 CA5521131 |
282 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773632625 CA5521133 |
283 | L>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 284 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377040065 rs1315055590 |
286 | D>G | No |
ClinGen gnomAD |
|
|
rs1013661646 CA208368722 |
286 | D>H | No |
ClinGen TOPMed |
|
|
rs375090685 CA208368725 |
289 | D>G | No |
ClinGen ESP |
|
|
rs1329525494 CA377040755 |
290 | L>I | No |
ClinGen gnomAD |
|
|
CA377040862 rs1236710841 |
294 | Q>* | No |
ClinGen gnomAD |
|
|
CA377040897 rs368002483 |
295 | A>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5521136 rs751811485 |
295 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs368002483 CA5521137 |
295 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5521139 rs753708973 |
296 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA377040928 rs1564884305 |
297 | F>S | No |
ClinGen Ensembl |
|
|
rs199983221 CA208368736 |
299 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
rs780983084 CA208368740 |
305 | D>E | No |
ClinGen Ensembl |
|
|
rs200902165 CA208368745 |
308 | P>A | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 315 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1235452860 CA377043976 |
316 | I>V | No |
ClinGen TOPMed |
|
|
rs1589080784 CA377043990 |
317 | Y>D | No |
ClinGen Ensembl |
|
| TCGA novel | 318 | P>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5521175 rs772390347 |
320 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA5521177 rs532201569 |
322 | Q>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA208376169 rs200987359 |
326 | C>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1410067243 CA377044172 |
328 | K>R | No |
ClinGen gnomAD |
|
|
CA208376173 rs552023236 |
330 | I>V | No |
ClinGen 1000Genomes |
|
|
rs201340003 CA208376177 |
331 | A>T | No |
ClinGen Ensembl |
|
|
CA377044223 rs1157736577 |
332 | L>W | No |
ClinGen TOPMed gnomAD |
|
|
rs200610338 CA5521181 |
333 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs751410492 CA5521182 |
335 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM1348653 rs201583982 CA208376193 |
341 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA5521183 rs147909071 |
341 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5521184 rs767148239 |
342 | N>T | No |
ClinGen ExAC gnomAD |
|
|
CA377044393 rs1292350491 |
343 | Y>C | No |
ClinGen gnomAD |
|
|
CA377044573 rs200058231 |
353 | V>F | No |
ClinGen TOPMed gnomAD |
|
|
rs200058231 CA208376210 |
353 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
rs777323664 CA5521187 |
354 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA208376216 rs141528984 |
354 | A>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA5521189 rs757804740 |
355 | G>E | No |
ClinGen ExAC gnomAD |
|
|
rs1017283750 CA208376232 |
356 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
rs116374368 CA5521190 |
357 | Q>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1480935024 CA377044619 |
358 | R>K | No |
ClinGen gnomAD |
|
|
rs1235876376 CA377044658 |
361 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1000020014 CA208382125 COSM194884 |
369 | A>T | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated Ensembl NCI-TCGA |
|
CA377045416 rs1471211487 |
372 | L>V | No |
ClinGen gnomAD |
|
|
CA5521209 rs763566753 |
373 | I>S | No |
ClinGen ExAC gnomAD |
|
|
rs753398498 CA5521210 |
375 | K>E | No |
ClinGen ExAC |
|
|
rs756786753 CA5521211 |
379 | D>A | No |
ClinGen ExAC TOPMed |
|
|
rs779440453 CA5521212 |
380 | C>G | No |
ClinGen ExAC gnomAD |
|
|
rs779440453 CA377045471 |
380 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs1305054381 CA377045473 |
380 | C>S | No |
ClinGen TOPMed |
|
|
CA377045486 rs1446148922 |
382 | A>P | No |
ClinGen gnomAD |
|
|
CA377045491 rs750895479 |
383 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs750895479 CA5521213 |
383 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA5521214 rs758727461 |
385 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs17855430 CA208382150 |
386 | D>A | No |
ClinGen TOPMed |
|
|
CA208382152 rs17855430 |
386 | D>G | No |
ClinGen TOPMed |
|
| TCGA novel | 386 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs17855431 CA208382155 |
387 | I>L | No |
ClinGen ExAC gnomAD |
|
|
rs17855431 CA5521215 |
387 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1255852295 CA377045526 |
389 | N>D | No |
ClinGen gnomAD |
|
|
rs17855432 CA208382162 |
389 | N>S | No |
ClinGen Ensembl |
|
|
CA208382166 rs989153228 |
390 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs201152568 CA208382748 |
392 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1051881207 CA208382756 |
394 | R>* | No |
ClinGen TOPMed |
|
|
rs766737138 CA5521233 |
394 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| TCGA novel | 396 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA208382767 rs757269120 |
396 | P>L | No |
ClinGen Ensembl |
|
|
rs752020872 CA5521234 |
396 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA377045583 rs1200857950 |
397 | R>G | No |
ClinGen gnomAD |
|
| TCGA novel | 397 | R>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377045613 rs1477293278 |
399 | P>Q | No |
ClinGen gnomAD |
|
|
CA208382770 rs1047393773 |
399 | P>T | No |
ClinGen Ensembl |
|
|
rs755416990 CA5521235 |
400 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748394475 COSM919612 CA5521237 |
403 | P>L | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA377045653 rs748394475 |
403 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377045659 rs1460011282 |
404 | L>F | No |
ClinGen TOPMed |
|
|
CA208382775 rs757140711 |
406 | I>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA5521240 rs149206117 |
411 | I>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA377045753 rs1340780587 |
412 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA5521241 rs771950281 |
416 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs775426483 CA5521242 |
419 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775426483 CA377045834 |
419 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5521243 rs746762337 |
421 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA5521245 rs776212608 |
421 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs371326132 CA5521244 |
421 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs201635394 CA5521247 |
425 | A>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs200690371 CA377045902 |
425 | A>P | No |
ClinGen TOPMed |
|
|
CA208382810 rs200690371 |
425 | A>T | No |
ClinGen TOPMed |
|
|
CA377045942 rs1362106598 |
428 | Y>C | No |
ClinGen gnomAD |
|
|
CA5521248 rs772640272 |
430 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1337495965 CA377045981 |
431 | D>G | No |
ClinGen TOPMed |
|
|
rs763496433 CA5521249 |
432 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs745364339 CA208382815 |
433 | V>I | No |
ClinGen gnomAD |
|
|
rs745364339 CA377045996 |
433 | V>L | No |
ClinGen gnomAD |
|
|
rs1184537805 CA377046010 |
434 | D>G | No |
ClinGen gnomAD |
|
|
CA208382832 rs199593180 |
435 | L>F | No |
ClinGen gnomAD |
|
|
CA5521250 rs766945174 |
435 | L>H | No |
ClinGen ExAC gnomAD |
|
|
rs199593180 CA377046018 |
435 | L>V | No |
ClinGen gnomAD |
|
|
rs755327185 CA5521252 |
436 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA5521253 rs768051584 |
437 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA377046036 rs1257460794 |
437 | I>V | No |
ClinGen TOPMed |
|
| TCGA novel | 439 | I>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1029118327 CA208382836 |
440 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs752958196 CA5521254 |
441 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs756329197 CA5521255 |
444 | K>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 446 | R>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1483205269 CA377046143 |
447 | P>L | No |
ClinGen gnomAD |
|
|
CA377046182 rs1325855305 |
451 | I>M | No |
ClinGen TOPMed |
|
|
CA208382849 rs267602551 |
452 | P>S | No |
ClinGen Ensembl |
|
|
CA377046194 rs1375598245 |
453 | S>G | No |
ClinGen gnomAD |
|
|
rs1200157325 CA377047031 |
455 | I>L | No |
ClinGen gnomAD |
|
|
rs200021101 CA208384424 |
455 | I>M | No |
ClinGen Ensembl |
|
|
rs201348222 CA208384427 |
456 | P>A | No |
ClinGen Ensembl |
|
|
CA208384434 rs146837595 |
457 | H>R | No |
ClinGen ESP |
|
|
CA208384439 rs140677498 |
462 | I>K | No |
ClinGen ESP |
|
|
rs1236259317 CA377047143 |
462 | I>M | No |
ClinGen Ensembl |
|
|
CA5521274 rs761122062 |
463 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA5521275 rs764179598 |
466 | R>K | No |
ClinGen ExAC gnomAD |
|
|
CA377047213 rs1406225734 |
467 | E>D | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 468 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA208384451 rs777705431 |
468 | P>R | No |
ClinGen Ensembl |
|
|
CA377047252 rs201948258 |
470 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201948258 CA5521277 |
470 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779026786 CA5521278 |
471 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA5521279 rs751498023 |
473 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754951946 CA5521280 |
479 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs145326137 CA5521281 |
483 | D>N | No |
ClinGen ESP ExAC gnomAD |
|
|
CA208384470 rs1063111 VAR_051976 |
484 | V>D | No |
ClinGen UniProt Ensembl dbSNP |
|
|
CA377047440 rs1312903591 |
484 | V>I | No |
ClinGen gnomAD |
|
|
CA208384476 rs1063112 |
485 | I>T | No |
ClinGen Ensembl |
|
| TCGA novel | 486 | I>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA208384488 rs771126114 |
488 | E>Q | No |
ClinGen gnomAD |
|
|
rs1063114 CA208384494 |
490 | C>* | No |
ClinGen Ensembl |
|
|
rs369274325 CA5521285 |
495 | G>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 497 | Y>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 498 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1173354902 CA377047662 |
499 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
CA5521287 rs549636735 |
499 | K>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA5521288 rs549636735 |
499 | K>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 500 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5521289 rs772656917 |
501 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs202116390 CA208384513 |
502 | C>R | No |
ClinGen gnomAD |
|
|
rs150099719 CA5521290 |
502 | C>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs201090108 CA5521291 |
503 | N>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs116499760 CA5521292 |
504 | P>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs909912033 CA208384542 |
505 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA377047809 rs1386664764 |
510 | I>V | No |
ClinGen gnomAD |
|
|
rs750553699 CA5521296 |
511 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs1285575541 CA377047864 |
514 | P>R | No |
ClinGen gnomAD |
|
| TCGA novel | 515 | P>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA208384560 rs201863201 |
515 | P>L | No |
ClinGen Ensembl |
|
|
rs1224464761 CA377047882 |
516 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA377047885 COSM277278 rs1247003248 |
516 | R>Q | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs201058854 CA5521300 |
518 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377047921 rs1397000657 |
519 | K>E | No |
ClinGen TOPMed |
|
|
rs200415719 CA208384571 |
519 | K>N | No |
ClinGen 1000Genomes TOPMed |
|
| TCGA novel | 520 | E>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5521301 rs61754500 |
522 | A>V | No |
ClinGen TOPMed |
|
|
rs1208799614 CA377047980 |
523 | Y>C | No |
ClinGen gnomAD |
|
|
rs199649997 CA208384586 |
525 | S>L | No |
ClinGen Ensembl |
|
|
rs748917510 CA5521304 |
527 | L>W | No |
ClinGen ExAC gnomAD |
|
|
rs770755510 CA377048049 |
528 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA5521305 rs770755510 |
528 | P>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1450189602 CA377048096 |
532 | L>F | No |
ClinGen gnomAD |
|
|
rs1292260091 CA377048100 |
532 | L>R | No |
ClinGen gnomAD |
|
|
CA377048119 rs1224283349 |
534 | V>I | No |
ClinGen gnomAD |
|
| TCGA novel | 535 | S>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5521308 rs116040871 RCV000948374 |
536 | E>K | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA377048162 rs1308551562 |
537 | D>N | No |
ClinGen gnomAD |
|
|
CA5521309 rs199497583 |
539 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA208384620 rs199497583 |
539 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5521310 rs192990424 |
539 | S>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA5521311 rs768902657 |
541 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA208384641 rs892821661 |
542 | E>V | No |
ClinGen TOPMed |
|
|
CA377048247 rs1357873484 |
543 | R>G | No |
ClinGen TOPMed |
|
|
CA377048253 rs1320870381 |
543 | R>T | No |
ClinGen gnomAD |
|
|
rs777098039 CA5521312 |
544 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA377048290 rs1475785988 |
546 | P>A | No |
ClinGen gnomAD |
|
|
rs201723648 CA208384651 |
547 | P>S | No |
ClinGen Ensembl |
|
|
rs1007238332 CA208384654 |
548 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA377048315 rs1307916842 |
548 | D>H | No |
ClinGen TOPMed |
|
| TCGA novel | 548 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs373331174 COSM377396 CA208384663 |
549 | S>C | lung [Cosmic] | No |
ClinGen cosmic curated ESP TOPMed |
|
CA377048360 rs1235780462 |
551 | V>A | No |
ClinGen gnomAD |
|
|
CA208384667 rs758444346 |
551 | V>M | No |
ClinGen Ensembl |
|
|
CA377048364 rs1446102743 |
552 | I>V | No |
ClinGen gnomAD |
|
|
CA5521315 rs773456695 |
553 | V>F | No |
ClinGen ExAC gnomAD |
|
|
rs377735046 CA208384674 |
556 | L>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs919162147 CA377048422 |
557 | D>H | No |
ClinGen gnomAD |
|
|
CA208384695 rs919162147 |
557 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1330804026 CA377048462 |
559 | A>G | No |
ClinGen gnomAD |
|
| TCGA novel | 559 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1051041309 CA208384703 |
559 | A>T | No |
ClinGen Ensembl |
|
|
CA5521316 rs762955289 |
560 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs756116247 CA5521319 |
564 | D>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 564 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs763773932 CA5521320 |
565 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA208384716 rs942784892 |
566 | L>S | No |
ClinGen Ensembl |
|
|
CA5521323 rs144625497 |
573 | G>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs745527350 CA5521324 |
574 | C>G | No |
ClinGen ExAC gnomAD |
|
|
rs1462243145 CA377048718 |
574 | C>Y | No |
ClinGen gnomAD |
|
|
rs757828571 CA5521325 |
575 | M>K | No |
ClinGen ExAC gnomAD |
|
|
rs779678154 CA5521326 |
576 | E>K | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 577 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs747447296 CA5521327 |
578 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA377048872 rs1589084920 |
580 | Q>R | No |
ClinGen Ensembl |
|
| TCGA novel | 581 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377048928 rs769257926 |
583 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377048939 rs777147954 |
583 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA5521329 rs769257926 |
583 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA208384756 rs201096600 |
583 | Q>P | No |
ClinGen Ensembl |
|
|
rs748473217 CA5521332 |
585 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs202044007 CA208384771 |
586 | R>G | No |
ClinGen Ensembl |
|
|
CA5521334 rs773365044 |
586 | R>T | No |
ClinGen ExAC gnomAD |
|
|
rs763220339 CA5521335 |
587 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA377049064 rs1199455034 |
589 | E>G | No |
ClinGen TOPMed |
|
|
rs1387799393 CA377049080 |
590 | S>R | No |
ClinGen gnomAD |
|
|
rs370128548 CA208384783 |
591 | I>V | No |
ClinGen ESP TOPMed |
|
|
CA5521338 rs759555466 |
594 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs139635382 CA5521339 |
595 | M>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA208384786 rs1039769206 |
595 | M>V | No |
ClinGen Ensembl |
|
|
CA5521340 rs753828684 |
597 | N>D | No |
ClinGen ExAC gnomAD |
|
|
CA5521342 rs765178020 |
601 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA5521343 rs750092788 |
603 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA208384811 rs533321736 |
604 | G>D | No |
ClinGen TOPMed |
|
|
CA5521344 rs200296961 |
604 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377049402 rs533321736 |
604 | G>V | No |
ClinGen TOPMed |
|
|
CA5521345 rs779413432 |
607 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs963095350 CA208384819 |
611 | N>H | No |
ClinGen TOPMed |
|
|
CA377049551 rs1193665057 |
612 | E>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 614 | T>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377049615 rs1414023245 |
615 | S>A | No |
ClinGen TOPMed |
|
|
rs781614748 CA5521348 |
616 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs991510115 CA208384822 |
620 | V>A | No |
ClinGen TOPMed |
|
|
rs991510115 CA377049718 |
620 | V>G | No |
ClinGen TOPMed |
|
| TCGA novel | 621 | R>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377049751 rs1160645506 |
623 | C>S | No |
ClinGen gnomAD |
|
| TCGA novel | 624 | W>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377049824 rs567829185 |
628 | V>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs567829185 CA5521350 |
628 | V>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs199770148 CA5521351 |
630 | K>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1296052737 CA377049872 |
631 | E>D | No |
ClinGen gnomAD |
|
|
rs1451930340 CA377049901 |
633 | I>M | No |
ClinGen TOPMed |
|
|
rs1332764907 CA377049909 |
634 | S>N | No |
ClinGen gnomAD |
|
|
CA377049932 rs1410882240 |
635 | R>S | No |
ClinGen TOPMed |
|
|
rs771135356 CA5521353 |
636 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5521352 rs201647881 |
636 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1342236378 CA377049942 |
637 | L>I | No |
ClinGen gnomAD |
|
|
rs1255389662 CA377049960 |
638 | D>G | No |
ClinGen TOPMed |
|
|
rs1238826257 CA377051762 |
640 | N>S | No |
ClinGen gnomAD |
|
|
rs766102594 CA5521379 |
641 | Q>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 645 | L>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA208386494 rs918503045 |
646 | P>L | No |
ClinGen TOPMed |
|
|
CA377051891 rs1351830584 |
646 | P>T | No |
ClinGen TOPMed |
|
|
rs1246391112 COSM215429 CA377051955 |
649 | R>C | Variant assessed as Somatic; impact. endometrium central_nervous_system [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
rs1162652974 CA594255386 |
650 | Y>* | No |
ClinGen gnomAD |
|
|
rs752153256 CA5521383 |
650 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377051986 rs1332048288 |
651 | I>V | No |
ClinGen TOPMed |
|
|
rs1410863068 CA377052101 |
656 | E>K | No |
ClinGen gnomAD |
|
|
CA208386500 rs199502996 |
657 | V>L | No |
ClinGen Ensembl |
|
|
CA208386502 rs984395300 |
658 | Y>H | No |
ClinGen TOPMed |
|
|
rs778184510 CA5521385 |
660 | D>A | No |
ClinGen ExAC gnomAD |
|
|
CA5521387 rs757624637 |
661 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA5521386 rs754313614 |
661 | S>P | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 661 | S>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 662 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 663 | D>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1246002188 CA377052309 |
665 | V>D | No |
ClinGen gnomAD |
|
|
CA5521389 rs201730062 |
665 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA377052324 rs1379252260 |
666 | L>S | No |
ClinGen TOPMed |
|
|
rs772469360 CA5521390 |
667 | S>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs780449017 CA5521391 |
667 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs772469360 CA208386515 |
667 | S>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA377052342 rs1258853759 |
668 | S>P | No |
ClinGen gnomAD |
|
|
rs768864413 CA5521394 |
670 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA5521393 rs768864413 |
670 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
rs114575266 CA5521396 |
671 | C>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs377449611 CA5521397 |
673 | S>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA377052432 rs1589087439 |
673 | S>R | No |
ClinGen Ensembl |
|
|
CA377052430 rs1186590766 |
673 | S>T | No |
ClinGen gnomAD |
|
|
rs759347614 CA5521398 |
675 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA5521399 rs759347614 |
675 | S>G | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1312104590 CA377052483 |
676 | D>G | No |
ClinGen Ensembl |
|
|
rs1589087464 CA377052585 |
681 | Q>* | No |
ClinGen Ensembl |
|
|
COSM1474709 CA377052594 rs1422369208 |
681 | Q>H | breast [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA377052587 rs1171276858 |
681 | Q>P | No |
ClinGen gnomAD |
|
|
rs1055202028 CA208386544 |
683 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA5521402 rs114572830 |
684 | S>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs114572830 CA208386547 |
684 | S>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs754362414 CA5521403 |
685 | L>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754362414 CA377052650 |
685 | L>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5521405 rs765609303 |
688 | P>H | No |
ClinGen ExAC gnomAD |
|
|
CA5521404 rs757740493 |
688 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA377052704 rs1223523187 |
689 | M>L | No |
ClinGen TOPMed |
|
|
CA5521406 rs750707792 |
689 | M>R | No |
ClinGen ExAC gnomAD |
|
|
rs1223523187 CA377052701 |
689 | M>V | No |
ClinGen TOPMed |
|
|
CA377052759 rs1279371482 |
693 | S>N | No |
ClinGen gnomAD |
|
|
rs140030776 CA5521407 |
695 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1287332193 CA377052783 |
696 | E>G | No |
ClinGen gnomAD |
|
| TCGA novel | 697 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA377052787 rs1225337110 |
697 | E>Q | No |
ClinGen gnomAD |
|
|
CA377052795 rs1282280106 |
698 | F>L | No |
ClinGen TOPMed |
|
|
rs1243937141 CA377052815 |
699 | Y>C | No |
ClinGen TOPMed |
|
|
CA5521409 rs747251569 |
700 | N>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA208386555 rs116459300 |
700 | N>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA5521410 rs116459300 |
700 | N>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA377052842 rs1255205628 |
701 | G>D | No |
ClinGen gnomAD |
|
|
CA5521411 rs201327262 CA377052876 |
703 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs151026272 CA208386559 |
704 | D>A | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs778261267 CA208386562 |
705 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs749218988 CA5521412 |
706 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA208386572 rs868057033 |
707 | D>E | No |
ClinGen Ensembl |
|
|
rs1007564772 CA208386567 |
707 | D>H | No |
ClinGen Ensembl |
|
|
rs1476767773 CA377052939 |
708 | V>A | No |
ClinGen gnomAD |
|
|
CA377052950 rs1377658006 |
709 | P>R | No |
ClinGen gnomAD |
|
|
rs770824011 CA377052945 |
709 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs770824011 CA5521413 |
709 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA377052955 rs1465268496 |
710 | E>Q | No |
ClinGen gnomAD |
|
|
CA5521414 rs774376548 |
711 | R>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5521415 rs114182972 |
712 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA377052999 rs771866041 |
713 | G>* | No |
ClinGen ExAC gnomAD |
|
|
rs771866041 CA5521416 |
713 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA377053008 rs1374102040 |
714 | G>R | No |
ClinGen TOPMed |
|
|
CA5521417 rs775109357 |
723 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA377053123 rs1299110584 |
723 | D>H | No |
ClinGen gnomAD |
|
|
rs201854199 CA5521418 |
724 | Q>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA377053141 rs1195962687 |
724 | Q>P | No |
ClinGen TOPMed |
|
|
rs201112743 CA208386590 |
725 | E>K | No |
ClinGen TOPMed |
|
|
CA5521420 rs776051187 |
726 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781343147 CA5521421 |
727 | I>SVN* | No |
ClinGen ExAC |
|
|
rs202021325 CA208386596 |
727 | I>T | No |
ClinGen Ensembl |
|
|
CA5521423 rs761406151 |
728 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5521422 rs761406151 |
728 | N>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs977473225 CA208386600 |
729 | E>D | No |
ClinGen gnomAD |
|
|
rs35224060 CA377053235 |
731 | I>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA5521425 rs758781892 |
731 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA5521424 rs35224060 |
731 | I>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA377053257 rs1486696314 |
732 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
rs766908589 CA5521426 |
733 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs1564896279 CA377053305 |
736 | E>K | No |
ClinGen Ensembl |
|
|
CA377053347 rs1252932360 |
739 | D>Y | No |
ClinGen TOPMed |
|
|
rs1188847251 CA377053384 |
742 | Y>* | No |
ClinGen gnomAD |
|
| TCGA novel | 745 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs756220511 CA5521431 |
745 | N>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA |
|
CA377053453 rs1471466430 |
747 | S>* | No |
ClinGen gnomAD |
No associated diseases with Q96EB6
1 regional properties for Q96EB6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Sirtuin family, catalytic core domain | 236 - 496 | IPR026590 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.1.286 | Transferring groups other than amino-acyl groups |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
16 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| chromatin silencing complex | Any protein complex that mediates changes in chromatin structure that result in transcriptional silencing. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| eNoSc complex | A chromatin silencing complex that recruits histone-modifying enzymes and upregulates silencing of rDNA in response to glucose starvation. |
| euchromatin | A dispersed and relatively uncompacted form of chromatin that is in a transcription-competent conformation. |
| fibrillar center | A structure found most metazoan nucleoli, but not usually found in lower eukaryotes; surrounded by the dense fibrillar component; the zone of transcription from multiple copies of the pre-rRNA genes is in the border region between these two structures. |
| heterochromatin | A compact and highly condensed form of chromatin that is refractory to transcription. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| nuclear envelope | The double lipid bilayer enclosing the nucleus and separating its contents from the rest of the cytoplasm; includes the intermembrane space, a gap of width 20-40 nm (also called the perinuclear space). |
| nuclear inner membrane | The inner, i.e. lumen-facing, lipid bilayer of the nuclear envelope. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| PML body | A class of nuclear body; they react against SP100 auto-antibodies (PML, promyelocytic leukemia); cells typically contain 10-30 PML bodies per nucleus; alterations in the localization of PML bodies occurs after viral infection. |
| rDNA heterochromatin | A region of heterochromatin located at the rDNA repeats in a chromosome. |
25 GO annotations of molecular function
| Name | Definition |
|---|---|
| bHLH transcription factor binding | Binding to a basic Helix-Loop-Helix (bHLH) superfamily of transcription factors, important regulatory components in transcriptional networks of many developmental pathways. |
| deacetylase activity | Catalysis of the hydrolysis of an acetyl group or groups from a substrate molecule. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| histone binding | Binding to a histone, any of a group of water-soluble proteins found in association with the DNA of eukaryotic or archaeal chromosomes. They are involved in the condensation and coiling of chromosomes during cell division and have also been implicated in gene regulation and DNA replication. They may be chemically modified (methylated, acetlyated and others) to regulate gene transcription. |
| histone deacetylase activity | Catalysis of the reaction: histone N6-acetyl-L-lysine + H2O = histone L-lysine + acetate. This reaction represents the removal of an acetyl group from a histone, a class of proteins complexed to DNA in chromatin and chromosomes. |
| HLH domain binding | Binding to a Helix Loop Helix domain, a domain of 40-50 residues that occurs in specific DNA-binding proteins that act as transcription factors. The domain is formed of two amphipathic helices joined by a variable length linker region that can form a loop and it mediates protein dimerization. |
| identical protein binding | Binding to an identical protein or proteins. |
| keratin filament binding | Binding to a keratin filament, an intermediate filament composed of acidic and basic keratins (types I and II), typically expressed in epithelial cells. |
| metal ion binding | Binding to a metal ion. |
| mitogen-activated protein kinase binding | Binding to a mitogen-activated protein kinase. |
| NAD+ binding | Binding to the oxidized form, NAD, of nicotinamide adenine dinucleotide, a coenzyme involved in many redox and biosynthetic reactions. |
| NAD-dependent histone deacetylase activity | Catalysis of the reaction: histone N6-acetyl-L-lysine + H2O = histone L-lysine + acetate. This reaction requires the presence of NAD, and represents the removal of an acetyl group from a histone. |
| NAD-dependent histone deacetylase activity (H3-K9 specific) | Catalysis of the reaction: histone H3 N6-acetyl-L-lysine (position 9) + H2O = histone H3 L-lysine (position 9) + acetate. This reaction requires the presence of NAD, and represents the removal of an acetyl group from lysine at position 9 of the histone H3 protein. |
| NAD-dependent histone decrotonylase activity | Catalysis of the reaction: H2O + N6-(2E)-butenoyl-L-lysyl- + NAD+ = 2''-O-(2E)-but-2-enoyl-ADP-D-ribose + L-lysyl- |
| NAD-dependent protein deacetylase activity | Catalysis of the removal of one or more acetyl groups from a protein, requiring NAD. |
| nuclear receptor binding | Binding to a nuclear receptor protein. Nuclear receptor proteins are DNA-binding transcription factors which are regulated by binding to a ligand. |
| p53 binding | Binding to one of the p53 family of proteins. |
| promoter-specific chromatin binding | Binding to a section of chromatin that is associated with gene promoter sequences of DNA. |
| protein C-terminus binding | Binding to a protein C-terminus, the end of a peptide chain at which the 1-carboxyl function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| protein lysine deacetylase activity | Catalysis of the reaction: H2O + N6-acetyl-L-lysyl- |
| protein-propionyllysine depropionylase activity | Catalysis of the reaction:H2O + N(6)-propanoyl-L-lysyl- + NAD(+) = 3''-O-propanoyl-ADP-D-ribose + L-lysyl- |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| transcription coactivator activity | A transcription coregulator activity that activates or increases the transcription of specific gene sets via binding to a DNA-bound DNA-binding transcription factor, either on its own or as part of a complex. Coactivators often act by altering chromatin structure and modifications. For example, one class of transcription coactivators modifies chromatin structure through covalent modification of histones. A second class remodels the conformation of chromatin in an ATP-dependent fashion. A third class modulates interactions of DNA-bound DNA-binding transcription factors with other transcription coregulators. A fourth class of coactivator activity is the bridging of a DNA-binding transcription factor to the general (basal) transcription machinery. The Mediator complex, which bridges sequence-specific DNA binding transcription factors and RNA polymerase, is also a transcription coactivator. |
| transcription corepressor activity | A transcription coregulator activity that represses or decreases the transcription of specific gene sets via binding to a DNA-bound DNA-binding transcription factor, either on its own or as part of a complex. Corepressors often act by altering chromatin structure and modifications. For example, one class of transcription corepressors modifies chromatin structure through covalent modification of histones. A second class remodels the conformation of chromatin in an ATP-dependent fashion. A third class modulates interactions of DNA-bound DNA-binding transcription factors with other transcription coregulators. |
116 GO annotations of biological process
| Name | Definition |
|---|---|
| angiogenesis | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels. |
| behavioral response to starvation | Any process that results in a change in the behavior of an organism as a result of deprivation of nourishment. |
| cellular glucose homeostasis | A cellular homeostatic process involved in the maintenance of an internal steady state of glucose within a cell or between a cell and its external environment. |
| cellular response to DNA damage stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism. |
| cellular response to glucose starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of glucose. |
| cellular response to hydrogen peroxide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hydrogen peroxide (H2O2) stimulus. |
| cellular response to hypoxia | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension. Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%, results in metabolic adaptation at both the cellular and organismal level. |
| cellular response to ionizing radiation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a ionizing radiation stimulus. Ionizing radiation is radiation with sufficient energy to remove electrons from atoms and may arise from spontaneous decay of unstable isotopes, resulting in alpha and beta particles and gamma rays. Ionizing radiation also includes X-rays. |
| cellular response to leukemia inhibitory factor | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a leukemia inhibitory factor stimulus. |
| cellular response to starvation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of nourishment. |
| cellular response to tumor necrosis factor | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a tumor necrosis factor stimulus. |
| cellular triglyceride homeostasis | Any process involved in the maintenance of an internal steady state of triglyceride within a cell or between a cell and its external environment. |
| cholesterol homeostasis | Any process involved in the maintenance of an internal steady state of cholesterol within an organism or cell. |
| chromatin organization | The assembly or remodeling of chromatin composed of DNA complexed with histones, other associated proteins, and sometimes RNA. |
| circadian regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression such that an expression pattern recurs with a regularity of approximately 24 hours. |
| DNA methylation-dependent heterochromatin assembly | Repression of transcription by methylation of DNA, leading to the formation of heterochromatin. |
| DNA synthesis involved in DNA repair | Synthesis of DNA that proceeds from the broken 3' single-strand DNA end and uses the homologous intact duplex as the template. |
| energy homeostasis | Any process involved in the balance between food intake (energy input) and energy expenditure. |
| fatty acid homeostasis | Any process involved in the maintenance of an internal steady state of fatty acid within an organism or cell. |
| heterochromatin assembly | An epigenetic gene silencing mechanism in which chromatin is compacted into heterochromatin, resulting in a chromatin conformation refractory to transcription. This process starts with heterochromatin nucleation, its spreading, and ends with heterochromatin boundary formation. |
| histone deacetylation | The modification of histones by removal of acetyl groups. |
| histone H3 deacetylation | The modification of histone H3 by the removal of one or more acetyl groups. |
| intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | The series of molecular signals in which an intracellular signal is conveyed to trigger the apoptotic death of a cell. The pathway is induced by the cell cycle regulator phosphoprotein p53, or an equivalent protein, in response to the detection of DNA damage, and ends when the execution phase of apoptosis is triggered. |
| leptin-mediated signaling pathway | The series of molecular signals initiated by leptin binding to its receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. Leptin is a hormone manufactured primarily in the adipocytes of white adipose tissue, and the level of circulating leptin is directly proportional to the total amount of fat in the body. |
| macrophage differentiation | The process in which a relatively unspecialized monocyte acquires the specialized features of a macrophage. |
| muscle organ development | The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work. |
| negative regulation of androgen receptor signaling pathway | Any process that decreases the rate, frequency, or extent of the androgen receptor signaling pathway. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of cAMP-dependent protein kinase activity | Any process that stops, prevents or reduces the frequency, rate or extent of cAMP-dependent protein kinase activity. |
| negative regulation of cell cycle | Any process that stops, prevents or reduces the rate or extent of progression through the cell cycle. |
| negative regulation of cell growth | Any process that stops, prevents, or reduces the frequency, rate, extent or direction of cell growth. |
| negative regulation of cellular response to testosterone stimulus | Any process that stops, prevents or reduces the frequency, rate or extent of cellular response to testosterone stimulus. |
| negative regulation of cellular senescence | Any process that stops, prevents or reduces the frequency, rate or extent of cellular senescence. |
| negative regulation of DNA damage response, signal transduction by p53 class mediator | Any process that stops, prevents, or reduces the frequency, rate or extent of the cascade of processes induced by the cell cycle regulator phosphoprotein p53, or an equivalent protein, in response to the detection of DNA damage. |
| negative regulation of DNA-binding transcription factor activity | Any process that stops, prevents, or reduces the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
| negative regulation of DNA-templated transcription | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| negative regulation of fat cell differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of adipocyte differentiation. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| negative regulation of helicase activity | Any process that stops or reduces the activity of a helicase. |
| negative regulation of histone H3-K14 acetylation | Any process that stops, prevents, or reduces the frequency, rate or extent of the addition of an acetyl group to histone H3 at position 14 of the histone. |
| negative regulation of histone H3-K9 trimethylation | Any process that stops, prevents or reduces the frequency, rate or extent of histone H3-K9 trimethylation. |
| negative regulation of histone H4-K16 acetylation | Any process that stops, prevents or reduces the frequency, rate or extent of histone H4-K16 acetylation. |
| negative regulation of I-kappaB kinase/NF-kappaB signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of -kappaB kinase/NF-kappaB signaling. |
| negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | Any process that stops, prevents or reduces the frequency, rate or extent of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator. |
| negative regulation of neuron death | Any process that stops, prevents or reduces the frequency, rate or extent of neuron death. |
| negative regulation of NF-kappaB transcription factor activity | Any process that stops, prevents, or reduces the frequency, rate or extent of the activity of the transcription factor NF-kappaB. |
| negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of an oxidative stress-induced intrinsic apoptotic signaling pathway. |
| negative regulation of peptidyl-lysine acetylation | Any process that stops, prevents or reduces the frequency, rate or extent of peptidyl-lysine acetylation. |
| negative regulation of phosphorylation | Any process that stops, prevents or decreases the rate of addition of phosphate groups to a molecule. |
| negative regulation of prostaglandin biosynthetic process | Any process that stops, prevents, or reduces the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of prostaglandin. |
| negative regulation of protein acetylation | Any process that stops, prevents or reduces the frequency, rate or extent of protein acetylation. |
| negative regulation of protein kinase B signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| negative regulation of TOR signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of TOR signaling. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| negative regulation of transforming growth factor beta receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of any TGF-beta receptor signaling pathway. |
| ovulation from ovarian follicle | The process leading to the rupture of the follicle, releasing the centrally located oocyte into the oviduct. An example of this is found in Mus musculus. |
| peptidyl-lysine acetylation | The acetylation of peptidyl-lysine. |
| peptidyl-lysine deacetylation | The removal of an acetyl group from an acetylated lysine residue in a peptide or protein. |
| positive regulation of adaptive immune response | Any process that activates or increases the frequency, rate, or extent of an adaptive immune response. |
| positive regulation of adipose tissue development | Any process that activates or increases the frequency, rate or extent of adipose tissue development. |
| positive regulation of angiogenesis | Any process that activates or increases angiogenesis. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of blood vessel endothelial cell migration | Any process that activates or increases the frequency, rate or extent of the migration of the endothelial cells of blood vessels. |
| positive regulation of cAMP-dependent protein kinase activity | Any process that activates or increases the frequency, rate or extent of cAMP-dependent protein kinase activity. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of cellular senescence | Any process that activates or increases the frequency, rate or extent of cellular senescence. |
| positive regulation of cholesterol efflux | Any process that increases the frequency, rate or extent of cholesterol efflux. Cholesterol efflux is the directed movement of cholesterol, cholest-5-en-3-beta-ol, out of a cell or organelle. |
| positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that activates or increases the activity of a cysteine-type endopeptidase involved in the apoptotic process. |
| positive regulation of DNA repair | Any process that activates or increases the frequency, rate or extent of DNA repair. |
| positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | Any process that activates or increases the frequency, rate or extent of an endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway. |
| positive regulation of endothelial cell proliferation | Any process that activates or increases the rate or extent of endothelial cell proliferation. |
| positive regulation of gluconeogenesis | Any process that activates or increases the frequency, rate or extent of gluconeogenesis. |
| positive regulation of histone deacetylation | Any process that activates or increases the frequency, rate or extent of the removal of acetyl groups from histones. |
| positive regulation of histone H3-K9 methylation | Any process that activates or increases the frequency, rate or extent of the covalent addition of a methyl group to the lysine at position 9 of histone H3. |
| positive regulation of histone methylation | Any process that activates or increases the frequency, rate or extent of the covalent addition of methyl groups to histones. |
| positive regulation of insulin receptor signaling pathway | Any process that increases the frequency, rate or extent of insulin receptor signaling. |
| positive regulation of macroautophagy | Any process, such as recognition of nutrient depletion, that activates or increases the rate of macroautophagy to bring cytosolic macromolecules to the vacuole/lysosome for degradation. |
| positive regulation of macrophage apoptotic process | Any process that activates or increases the frequency, rate or extent of macrophage apoptotic process. |
| positive regulation of macrophage cytokine production | Any process that increases the rate, frequency or extent of macrophage cytokine production. Macrophage cytokine production is the appearance of a chemokine due to biosynthesis or secretion following a cellular stimulus, resulting in an increase in its intracellular or extracellular levels. |
| positive regulation of MHC class II biosynthetic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of MHC class II. |
| positive regulation of phosphatidylinositol 3-kinase signaling | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the phosphatidylinositol 3-kinase cascade. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| positive regulation of smooth muscle cell differentiation | Any process that activates or increases the frequency, rate or extent of smooth muscle cell differentiation. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| proteasome-mediated ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| protein deacetylation | The removal of an acetyl group from a protein amino acid. An acetyl group is CH3CO-, derived from acetic |
| protein depropionylation | The removal of a propionyl group from a residue in a peptide or protein. |
| protein destabilization | Any process that decreases the stability of a protein, making it more vulnerable to degradative processes or aggregation. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| pyrimidine dimer repair by nucleotide-excision repair | The repair of UV-induced T-T, C-T, and C-C dimers by the recognition and removal of the damaged DNA strand from the DNA helix as an oligonucleotide. The small gap left in the DNA helix is filled in by the sequential action of DNA polymerase and DNA ligase. |
| rDNA heterochromatin assembly | The formation of heterochromatin at ribosomal DNA, characterized by the modified histone H3K9me3. |
| regulation of apoptotic process | Any process that modulates the occurrence or rate of cell death by apoptotic process. |
| regulation of bile acid biosynthetic process | Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of bile acids. |
| regulation of brown fat cell differentiation | Any process that modulates the rate, frequency, or extent of brown fat cell differentiation. Brown fat cell differentiation is the process in which a relatively unspecialized cell acquires specialized features of a brown adipocyte, an animal connective tissue cell involved in adaptive thermogenesis. Brown adipocytes contain multiple small droplets of triglycerides and a high number of mitochondria. |
| regulation of cell population proliferation | Any process that modulates the frequency, rate or extent of cell proliferation. |
| regulation of cellular response to heat | Any process that modulates the frequency, rate or extent of cellular response to heat. |
| regulation of centrosome duplication | Any process that modulates the frequency, rate or extent of centrosome duplication. Centrosome duplication is the replication of a centrosome, a structure comprised of a pair of centrioles and peri-centriolar material from which a microtubule spindle apparatus is organized. |
| regulation of endodeoxyribonuclease activity | Any process that modulates the frequency, rate or extent of endodeoxyribonuclease activity, the hydrolysis of ester linkages within deoxyribonucleic acid by creating internal breaks. |
| regulation of glucose metabolic process | Any process that modulates the rate, frequency or extent of glucose metabolism. Glucose metabolic processes are the chemical reactions and pathways involving glucose, the aldohexose gluco-hexose. |
| regulation of lipid storage | Any process that modulates the rate, frequency or extent of lipid storage. Lipid storage is the accumulation and maintenance in cells or tissues of lipids, compounds soluble in organic solvents but insoluble or sparingly soluble in aqueous solvents. Lipid reserves can be accumulated during early developmental stages for mobilization and utilization at later stages of development. |
| regulation of mitotic cell cycle | Any process that modulates the rate or extent of progress through the mitotic cell cycle. |
| regulation of peroxisome proliferator activated receptor signaling pathway | Any process that modulates the frequency, rate or extent of the peroxisome proliferator activated receptor signaling pathway. |
| regulation of protein serine/threonine kinase activity | Any process that modulates the rate, frequency, or extent of protein serine/threonine kinase activity. |
| regulation of smooth muscle cell apoptotic process | Any process that modulates the frequency, rate, or extent of smooth muscle cell apoptotic process. |
| regulation of transcription by glucose | Any process involving glucose that modulates the frequency, rate or extent or transcription. |
| response to hydrogen peroxide | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hydrogen peroxide (H2O2) stimulus. |
| response to insulin | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| response to leptin | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a leptin stimulus. Leptin is a hormone manufactured primarily in the adipocytes of white adipose tissue, and the level of circulating leptin is directly proportional to the total amount of fat in the body. It plays a key role in regulating energy intake and energy expenditure, including appetite and metabolism]. |
| response to oxidative stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| single strand break repair | The repair of single strand breaks in DNA. Repair of such breaks is mediated by the same enzyme systems as are used in base excision repair. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
| stress-induced premature senescence | A cellular senescence process associated with the dismantling of a cell as a response to environmental factors such as hydrogen peroxide or X-rays. |
| transforming growth factor beta receptor signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a transforming growth factor beta receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| triglyceride mobilization | The release of triglycerides, any triester of glycerol, from storage within cells or tissues, making them available for metabolism. |
| UV-damage excision repair | A DNA repair process that is initiated by an endonuclease that introduces a single-strand incision immediately 5' of a UV-induced damage site. UV-damage excision repair acts on both cyclobutane pyrimidine dimers (CPDs) and pyrimidine-pyrimidone 6-4 photoproducts (6-4PPs). |
| white fat cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a white adipocyte, an animal connective tissue cell involved in energy storage. White adipocytes have cytoplasmic lipids arranged in a unique vacuole. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9VK34 | Sirt1 | NAD-dependent histone deacetylase sirtuin-1 | Drosophila melanogaster (Fruit fly) | PR |
| Q8IXJ6 | SIRT2 | NAD-dependent protein deacetylase sirtuin-2 | Homo sapiens (Human) | EV |
| Q8N6T7 | SIRT6 | NAD-dependent protein deacylase sirtuin-6 | Homo sapiens (Human) | PR |
| Q9NTG7 | SIRT3 | NAD-dependent protein deacetylase sirtuin-3, mitochondrial | Homo sapiens (Human) | SS |
| Q923E4 | Sirt1 | NAD-dependent protein deacetylase sirtuin-1 | Mus musculus (Mouse) | PR |
| Q68FX9 | Sirt5 | NAD-dependent protein deacylase sirtuin-5, mitochondrial | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MADEAALALQ | PGGSPSAAGA | DREAASSPAG | EPLRKRPRRD | GPGLERSPGE | PGGAAPEREV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PAAARGCPGA | AAAALWREAE | AEAAAAGGEQ | EAQATAAAGE | GDNGPGLQGP | SREPPLADNL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| YDEDDDDEGE | EEEEAAAAAI | GYRDNLLFGD | EIITNGFHSC | ESDEEDRASH | ASSSDWTPRP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RIGPYTFVQQ | HLMIGTDPRT | ILKDLLPETI | PPPELDDMTL | WQIVINILSE | PPKRKKRKDI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NTIEDAVKLL | QECKKIIVLT | GAGVSVSCGI | PDFRSRDGIY | ARLAVDFPDL | PDPQAMFDIE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YFRKDPRPFF | KFAKEIYPGQ | FQPSLCHKFI | ALSDKEGKLL | RNYTQNIDTL | EQVAGIQRII |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QCHGSFATAS | CLICKYKVDC | EAVRGDIFNQ | VVPRCPRCPA | DEPLAIMKPE | IVFFGENLPE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| QFHRAMKYDK | DEVDLLIVIG | SSLKVRPVAL | IPSSIPHEVP | QILINREPLP | HLHFDVELLG |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DCDVIINELC | HRLGGEYAKL | CCNPVKLSEI | TEKPPRTQKE | LAYLSELPPT | PLHVSEDSSS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| PERTSPPDSS | VIVTLLDQAA | KSNDDLDVSE | SKGCMEEKPQ | EVQTSRNVES | IAEQMENPDL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| KNVGSSTGEK | NERTSVAGTV | RKCWPNRVAK | EQISRRLDGN | QYLFLPPNRY | IFHGAEVYSD |
| 670 | 680 | 690 | 700 | 710 | 720 |
| SEDDVLSSSS | CGSNSDSGTC | QSPSLEEPME | DESEIEEFYN | GLEDEPDVPE | RAGGAGFGTD |
| 730 | 740 | ||||
| GDDQEAINEA | ISVKQEVTDM | NYPSNKS |