Q96BR1
Gene name |
SGK3 (CISK, SGKL) |
Protein name |
Serine/threonine-protein kinase Sgk3 |
Names |
EC 2.7.11.1 , Cytokine-independent survival kinase , Serum/glucocorticoid-regulated kinase 3 , Serum/glucocorticoid-regulated kinase-like |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:100533105, hsa:23678, |
EC number |
2.7.11.-: Protein-serine/threonine kinases |
Protein Class |
|
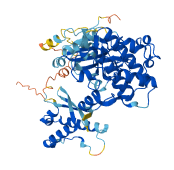
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
152-496 (Kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
303-326 (Activation loop from InterPro)
Target domain |
162-490 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Huang X et al. (2003) "Crystal structure of an inactive Akt2 kinase domain", Structure (London, England : 1993), 11, 21-30
- Truebestein L et al. (2021) "Structure of autoinhibited Akt1 reveals mechanism of PIP(3)-mediated activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
- Lučić I et al. (2018) "Conformational sampling of membranes by Akt controls its activation and inactivation", Proceedings of the National Academy of Sciences of the United States of America, 115, E3940-E3949
Autoinhibited structure

Activated structure

2 structures for Q96BR1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6EDX | X-ray | 201 A | A | 10-125 | PDB |
| AF-Q96BR1-F1 | Predicted | AlphaFoldDB |
407 variants for Q96BR1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs193920898 CA174151 RCV000149035 |
261 | H>R | Malignant tumor of prostate [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
| rs1253583314 | 2 | Q>E | No | gnomAD | |
| rs776685803 | 2 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs553948504 | 3 | R>G | No |
1000Genomes ExAC gnomAD |
|
| rs765398043 | 3 | R>K | No |
ExAC gnomAD |
|
| rs1807559581 | 5 | H>R | No | Ensembl | |
| rs765422599 | 6 | T>I | No | TOPMed | |
| rs1807559671 | 6 | T>S | No | Ensembl | |
| rs765422599 | 6 | T>S | No | TOPMed | |
| rs1807559936 | 7 | M>V | No | Ensembl | |
| rs1485724176 | 8 | D>E | No | gnomAD | |
| rs750459045 | 10 | K>E | No |
ExAC gnomAD |
|
| rs1427694029 | 11 | E>D | No | gnomAD | |
| rs1260637564 | 11 | E>K | No | gnomAD | |
| rs142472352 | 12 | S>G | No |
ESP ExAC gnomAD |
|
| rs1374592433 | 13 | C>Y | No | gnomAD | |
| rs766986095 | 14 | P>S | No |
ExAC gnomAD |
|
| rs1036217721 | 15 | S>N | No | TOPMed | |
| rs752166025 | 16 | V>I | No |
ExAC gnomAD |
|
| rs1807561283 | 17 | S>N | No |
TOPMed gnomAD |
|
|
rs1427992514 COSM3925476 |
21 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs146507110 | 22 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1298096883 | 23 | E>* | No | gnomAD | |
| rs1425598810 | 25 | R>T | No |
TOPMed gnomAD |
|
| rs1807562848 | 26 | E>K | No | TOPMed | |
| rs1281726110 | 30 | R>* | No | gnomAD | |
| rs1347927637 | 30 | R>G | No |
TOPMed gnomAD |
|
| rs777299349 | 30 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs777299349 | 30 | R>M | No |
ExAC TOPMed gnomAD |
|
| rs777299349 | 30 | R>T | No |
ExAC TOPMed gnomAD |
|
| rs1347927637 | 30 | R>W | No |
TOPMed gnomAD |
|
| rs1474371748 | 31 | F>L | No | TOPMed | |
| rs1186778201 | 31 | F>L | No | gnomAD | |
| TCGA novel | 35 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1298156892 | 39 | S>P | No | gnomAD | |
| rs959387847 | 43 | S>N | No | TOPMed | |
| rs1807797011 | 44 | E>* | No | TOPMed | |
| rs1807797011 | 44 | E>K | No | TOPMed | |
| rs1807797266 | 45 | W>* | No | TOPMed | |
| TCGA novel | 45 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763790706 | 47 | V>L | No |
ExAC gnomAD |
|
| rs756729751 | 48 | F>V | No |
ExAC gnomAD |
|
| rs922974165 | 49 | R>K | No | TOPMed | |
| rs778354101 | 50 | R>* | No |
ExAC gnomAD |
|
| rs866469551 | 51 | Y>C | No | TOPMed | |
| rs1283203345 | 51 | Y>H | No | Ensembl | |
| rs1469668623 | 52 | A>E | No | gnomAD | |
| rs745404100 | 53 | E>D | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 53 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1807798860 | 58 | Y>C | No | Ensembl | |
| rs1807798954 | 59 | N>D | No | gnomAD | |
| TCGA novel | 59 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs551009388 | 61 | L>* | No |
1000Genomes ExAC |
|
| rs779347222 | 62 | K>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 63 | K>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs149355530 | 64 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1563636611 | 64 | Q>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1357035961 | 67 | A>S | No |
TOPMed gnomAD |
|
| rs1357035961 | 67 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1290731101 | 68 | M>K | No |
TOPMed gnomAD |
|
| rs1808070515 | 68 | M>V | No | Ensembl | |
| rs1428226907 | 70 | L>V | No |
TOPMed gnomAD |
|
|
COSM1489381 COSM1489380 |
73 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1808070929 | 74 | A>S | No | Ensembl | |
| rs1362430508 | 75 | K>R | No |
TOPMed gnomAD |
|
| rs1278157968 | 76 | R>K | No |
TOPMed gnomAD |
|
| rs1278157968 | 76 | R>T | No |
TOPMed gnomAD |
|
| rs1375110081 | 77 | I>V | No |
TOPMed gnomAD |
|
| rs1347240660 | 78 | F>I | No | gnomAD | |
| rs1808071681 | 80 | D>N | No |
TOPMed gnomAD |
|
| TCGA novel | 82 | F>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1211762666 | 82 | F>L | No | gnomAD | |
| rs1282582183 | 84 | P>A | No | gnomAD | |
| rs1282582183 | 84 | P>S | No | gnomAD | |
| rs1445429064 | 85 | D>Y | No | gnomAD | |
| rs781283379 | 86 | F>V | No |
ExAC gnomAD |
|
| rs1808476573 | 87 | I>M | No | TOPMed | |
| rs752538144 | 88 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs756034666 | 91 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs779257049 | 91 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs746133153 | 92 | A>E | No |
ExAC gnomAD |
|
| rs746133153 | 92 | A>G | No |
ExAC gnomAD |
|
| rs1808477333 | 92 | A>T | No | TOPMed | |
| VAR_035636 | 92 | A>V | a breast cancer sample; somatic mutation [UniProt] | No | UniProt |
| rs1808477527 | 93 | G>R | No | gnomAD | |
| rs1488391217 | 96 | E>D | No | TOPMed | |
| rs772372105 | 96 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1279212836 | 97 | F>S | No |
TOPMed gnomAD |
|
| rs1355933890 | 98 | I>T | No | gnomAD | |
| rs1217359374 | 100 | N>D | No |
TOPMed gnomAD |
|
| rs1438707166 | 101 | L>V | No | Ensembl | |
| rs1346942385 | 102 | V>A | No |
TOPMed gnomAD |
|
| rs1056638872 | 103 | R>G | No | Ensembl | |
| rs1327900154 | 105 | P>S | No |
TOPMed gnomAD |
|
| rs1327900154 | 105 | P>T | No |
TOPMed gnomAD |
|
| rs917196477 | 106 | E>Q | No |
TOPMed gnomAD |
|
| rs1808479167 | 107 | L>F | No | Ensembl | |
| rs1238505623 | 108 | Y>H | No | gnomAD | |
| rs1238505623 | 108 | Y>N | No | gnomAD | |
| rs779950497 | 110 | H>N | No |
ExAC TOPMed gnomAD |
|
| rs1808875834 | 110 | H>Q | No | Ensembl | |
| rs779950497 | 110 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1363750277 | 111 | P>L | No |
TOPMed gnomAD |
|
| rs752734301 | 111 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1808876423 | 112 | D>G | No | Ensembl | |
| rs1053413373 | 112 | D>H | No | gnomAD | |
| rs990517924 | 113 | V>I | No | TOPMed | |
| rs201134569 | 114 | R>T | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3901076 COSM3901077 |
115 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763941506 | 115 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs753691709 | 115 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1307388997 | 118 | Q>L | No |
TOPMed gnomAD |
|
| rs758491010 | 119 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs1808878198 | 120 | D>G | No | Ensembl | |
| rs780326958 | 120 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs747055036 | 121 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1808878603 | 121 | S>N | No | Ensembl | |
| rs1585769606 | 122 | P>L | No | TOPMed | |
| rs1585769606 | 122 | P>R | No | TOPMed | |
| rs1461380916 | 122 | P>T | No | gnomAD | |
| rs1808879042 | 123 | K>E | No | Ensembl | |
| rs574203589 | 124 | H>L | No | gnomAD | |
| rs1168445012 | 124 | H>Y | No | gnomAD | |
| rs1407290946 | 125 | Q>P | No |
TOPMed gnomAD |
|
| rs1407290946 | 125 | Q>R | No |
TOPMed gnomAD |
|
| rs1403940747 | 126 | S>A | No | TOPMed | |
| rs1808879989 | 127 | D>E | No | gnomAD | |
| rs755143437 | 127 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs755143437 | 127 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1585769652 | 128 | P>A | No | TOPMed | |
| rs890597454 | 129 | S>T | No | Ensembl | |
| rs184264216 | 133 | D>G | No | 1000Genomes | |
| rs1356277366 | 133 | D>N | No | gnomAD | |
| TCGA novel | 135 | R>E | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1398014085 | 135 | R>G | No | gnomAD | |
| rs1293833639 | 135 | R>T | No | gnomAD | |
|
COSM1457883 COSM1457884 |
137 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1326233832 | 139 | K>E | No | gnomAD | |
| rs1326233832 | 139 | K>Q | No | gnomAD | |
| rs55878501 | 141 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781204914 | 141 | H>Y | No |
ExAC gnomAD |
|
|
COSM1489382 COSM1489383 |
142 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756552373 | 143 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs749604996 | 147 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs749604996 | 147 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs771380473 | 148 | N>S | No |
ExAC gnomAD |
|
| rs771380473 | 148 | N>T | No |
ExAC gnomAD |
|
| rs773848171 | 151 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1327216769 | 152 | S>C | No | gnomAD | |
| rs1809168719 | 152 | S>T | No |
TOPMed gnomAD |
|
| rs2130690304 | 153 | G>V | No | Ensembl | |
| rs1220517641 | 154 | N>I | No | gnomAD | |
| rs775084397 | 155 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs115712804 | 156 | H>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs115712804 | 156 | H>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1006341609 | 157 | A>G | No |
TOPMed gnomAD |
|
| rs1006341609 | 157 | A>V | No |
TOPMed gnomAD |
|
| rs779288300 | 162 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs200368158 | 163 | D>H | No | Ensembl | |
| rs771785674 | 164 | F>C | No |
ExAC TOPMed gnomAD |
|
| rs771785674 | 164 | F>Y | No |
ExAC TOPMed gnomAD |
|
| rs1809291865 | 167 | V>D | No | Ensembl | |
| rs779918683 | 167 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1809292106 | 168 | I>T | No | Ensembl | |
| COSM76405 | 169 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1353384001 | 169 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1809292400 | 172 | S>T | No | TOPMed | |
| rs776400322 | 175 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs752124707 | 176 | V>F | No |
ExAC gnomAD |
|
| rs752124707 | 176 | V>L | No |
ExAC gnomAD |
|
| rs749063520 | 177 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs749063520 | 177 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs770757970 | 179 | A>V | No |
ExAC gnomAD |
|
| rs774165780 | 180 | K>E | No |
ExAC gnomAD |
|
| rs764236858 | 181 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs760837872 COSM5026924 COSM5026925 |
181 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs372306972 | 182 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1172793059 | 183 | L>M | No | gnomAD | |
|
COSM486607 COSM486608 |
183 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1172793059 | 183 | L>V | No | gnomAD | |
| rs1809501653 | 184 | D>G | No | TOPMed | |
| rs1426898634 | 186 | K>E | No |
TOPMed gnomAD |
|
| rs2130709075 | 187 | F>C | No | Ensembl | |
| rs1216443244 | 187 | F>L | No | TOPMed | |
| rs761867027 | 187 | F>L | No |
ExAC gnomAD |
|
| rs1319600835 | 188 | Y>C | No | gnomAD | |
| rs1389138153 | 189 | A>V | No | gnomAD | |
| rs1809502954 | 192 | V>M | No | TOPMed | |
| rs1809503090 | 194 | Q>R | No | Ensembl | |
| rs1208907374 | 197 | I>M | No |
TOPMed gnomAD |
|
| rs1437354554 | 199 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1276706902 | 200 | N>S | No |
TOPMed gnomAD |
|
| rs766809256 | 201 | R>G | No |
ExAC gnomAD |
|
| rs1809504683 | 203 | E>K | No |
TOPMed gnomAD |
|
| rs761927874 | 204 | Q>R | No |
ExAC gnomAD |
|
| rs765445570 | 206 | H>R | No |
ExAC gnomAD |
|
| rs1357586907 | 208 | M>I | No | gnomAD | |
| rs773457219 | 208 | M>V | No |
ExAC TOPMed gnomAD |
|
|
COSM4850120 COSM4850121 |
210 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1472341296 | 211 | R>C | No |
TOPMed gnomAD |
|
| rs550671159 | 211 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA TOPMed gnomAD |
| rs1472341296 | 211 | R>S | No |
TOPMed gnomAD |
|
| rs763501787 | 212 | N>S | No |
ExAC gnomAD |
|
| rs767002581 | 213 | V>M | No |
ExAC gnomAD |
|
| rs1486698608 | 215 | L>M | No |
TOPMed gnomAD |
|
| rs781657155 | 218 | V>E | No | Ensembl | |
| rs538351516 | 219 | K>* | No | Ensembl | |
| rs373079510 | 220 | H>L | No |
ESP ExAC TOPMed |
|
| rs373079510 | 220 | H>R | No |
ESP ExAC TOPMed |
|
| rs1016260541 | 220 | H>Y | No |
TOPMed gnomAD |
|
| rs1181002886 | 221 | P>L | No | gnomAD | |
| rs1809513688 | 225 | G>A | No | TOPMed | |
| rs767178752 | 225 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1809514029 | 229 | S>C | No |
TOPMed gnomAD |
|
| rs376002305 | 232 | T>R | No |
ESP TOPMed |
|
| rs1350431820 | 233 | T>A | No | TOPMed | |
| rs139287562 | 234 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3942715 COSM3942716 |
234 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777221114 | 235 | K>T | No |
ExAC gnomAD |
|
| rs1417089611 | 238 | F>L | No |
TOPMed gnomAD |
|
| rs1429261706 | 240 | L>V | No | gnomAD | |
| rs757120302 | 244 | N>D | No |
ExAC gnomAD |
|
| rs1809516161 | 245 | G>A | No | TOPMed | |
| rs1809516315 | 246 | G>W | No | TOPMed | |
| rs1372577351 | 247 | E>G | No | gnomAD | |
|
TCGA novel rs1809735749 |
248 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1255118889 | 250 | F>S | No |
TOPMed gnomAD |
|
|
COSM1457892 COSM1457891 |
250 | F>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 251 | H>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4928296 COSM4928295 |
251 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753343248 | 252 | L>I | No |
ExAC gnomAD |
|
| rs778801923 | 253 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs778801923 | 253 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs750365774 | 254 | R>K | No |
ExAC gnomAD |
|
| rs758391298 | 255 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs758391298 | 255 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs913845064 | 256 | R>L | No |
TOPMed gnomAD |
|
| rs913845064 | 256 | R>Q | No |
TOPMed gnomAD |
|
| rs867803355 | 256 | R>W | No |
TOPMed gnomAD |
|
| rs143084838 | 257 | S>C | No |
1000Genomes ExAC gnomAD |
|
| rs770071015 | 259 | P>A | No |
ExAC gnomAD |
|
| rs749362374 | 262 | R>T | No |
ExAC gnomAD |
|
| rs1323181667 | 264 | R>G | No |
TOPMed gnomAD |
|
| rs868465468 | 264 | R>K | No | Ensembl | |
| rs771176965 | 267 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1809739651 | 270 | I>V | No | TOPMed | |
| rs1809739751 | 271 | A>G | No | TOPMed | |
| TCGA novel | 271 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1101125 COSM1101124 |
276 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775650552 | 280 | I>V | No |
ExAC gnomAD |
|
| rs1313011010 | 282 | I>T | No | Ensembl | |
| rs761095195 | 283 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1809740654 | 284 | Y>F | No | TOPMed | |
| rs1209273483 | 285 | R>G | No | gnomAD | |
| rs375908504 | 292 | I>N | No |
ESP ExAC gnomAD |
|
| rs375908504 | 292 | I>T | No |
ESP ExAC gnomAD |
|
|
COSM1101126 COSM1101127 |
293 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs747354281 | 296 | S>P | No |
ExAC gnomAD |
|
| rs769050993 | 297 | V>L | No |
ExAC gnomAD |
|
| rs1032147306 | 298 | G>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1809780958 | 300 | V>A | No | Ensembl | |
| rs1809780847 | 300 | V>I | No | gnomAD | |
| rs1236062277 | 303 | T>I | No | gnomAD | |
|
COSM750943 COSM750944 |
304 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758557227 | 314 | I>V | No |
ExAC gnomAD |
|
| rs780396359 | 316 | D>N | No |
ExAC gnomAD |
|
| rs1809781814 | 317 | T>N | No |
TOPMed gnomAD |
|
| rs1309516620 | 325 | P>A | No | gnomAD | |
| rs142484024 | 327 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1809874899 COSM6181151 COSM6181150 |
327 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic gnomAD |
| rs959482930 | 328 | L>F | No | Ensembl | |
| TCGA novel | 328 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs959482930 | 328 | L>V | No | Ensembl | |
| rs76769303 | 329 | A>S | No | Ensembl | |
| rs1809875560 | 330 | P>T | No | Ensembl | |
| rs1809875819 | 336 | Q>E | No | TOPMed | |
| rs1011486335 | 337 | P>H | No | Ensembl | |
| rs759490983 | 338 | Y>C | No |
ExAC gnomAD |
|
| rs1809876162 | 339 | D>N | No |
TOPMed gnomAD |
|
| rs1809876276 | 341 | T>A | No | Ensembl | |
| rs1244857651 | 341 | T>I | No | TOPMed | |
| rs1191859262 | 342 | V>I | No |
TOPMed gnomAD |
|
| rs1469906640 | 343 | D>G | No | Ensembl | |
| TCGA novel | 343 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760580623 | 344 | W>C | No |
ExAC TOPMed gnomAD |
|
| rs1169269391 | 346 | C>* | No | gnomAD | |
| rs1809877754 | 347 | L>F | No | TOPMed | |
| rs1176465510 | 349 | A>S | No |
TOPMed gnomAD |
|
| rs1176465510 | 349 | A>T | No |
TOPMed gnomAD |
|
| rs765358881 | 350 | V>I | No |
ExAC gnomAD |
|
| rs1327261422 | 352 | Y>F | No | TOPMed | |
| rs2130730737 | 352 | Y>H | No | Ensembl | |
| rs1286801966 | 354 | M>V | No |
TOPMed gnomAD |
|
|
rs750700898 VAR_041076 |
355 | L>P | No |
UniProt ExAC TOPMed dbSNP gnomAD |
|
| rs758582469 | 358 | L>S | No |
ExAC gnomAD |
|
|
COSM3650298 COSM3650297 |
359 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs761576561 | 359 | P>S | No |
ExAC gnomAD |
|
| TCGA novel | 360 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs766570570 | 362 | Y>C | No |
ExAC gnomAD |
|
| rs766570570 | 362 | Y>F | No |
ExAC gnomAD |
|
| rs1256492465 | 364 | R>* | No | gnomAD | |
| rs774604635 | 364 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs774604635 | 364 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM1101130 COSM1101131 |
365 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1027728991 | 366 | V>I | No |
TOPMed gnomAD |
|
| rs1027728991 | 366 | V>L | No |
TOPMed gnomAD |
|
| rs1810070768 | 367 | A>P | No | gnomAD | |
| rs767543430 | 369 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs952010662 | 372 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1289318995 | 376 | K>N | No | gnomAD | |
| rs1352765522 | 377 | P>H | No |
TOPMed gnomAD |
|
| rs1243212526 | 378 | L>I | No |
TOPMed gnomAD |
|
| rs1243212526 | 378 | L>V | No |
TOPMed gnomAD |
|
| rs1810072040 | 383 | G>R | No |
TOPMed gnomAD |
|
| rs756578171 | 385 | S>N | No |
ExAC gnomAD |
|
| rs764576243 | 386 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1810072531 | 387 | T>A | No | Ensembl | |
| rs1481842638 | 387 | T>I | No |
TOPMed gnomAD |
|
| rs1810072773 | 388 | A>T | No | TOPMed | |
| TCGA novel | 388 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1204064939 | 389 | W>* | No | gnomAD | |
| rs1204064939 | 389 | W>C | No | gnomAD | |
| rs2130739647 | 393 | E>D | No | Ensembl | |
| rs1810073303 | 397 | E>A | No | Ensembl | |
| rs754217242 | 399 | D>E | No |
ExAC gnomAD |
|
| rs1810073561 | 401 | Q>K | No | Ensembl | |
|
COSM1101133 rs1810073680 COSM1101132 |
403 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs1367767221 | 403 | R>Q | No |
TOPMed gnomAD |
|
| TCGA novel | 406 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1473086261 | 407 | K>R | No | gnomAD | |
| rs757750013 | 408 | E>G | No |
ExAC gnomAD |
|
| rs1279607748 | 409 | D>G | No | gnomAD | |
| rs146543614 | 409 | D>N | No | Ensembl | |
| rs1362386841 | 411 | L>P | No | gnomAD | |
| rs1810253006 | 412 | E>Q | No | TOPMed | |
| rs879156224 | 420 | E>* | No |
TOPMed gnomAD |
|
| rs2130748125 | 420 | E>G | No | Ensembl | |
| rs879156224 | 420 | E>K | No |
TOPMed gnomAD |
|
| rs879156224 | 420 | E>Q | No |
TOPMed gnomAD |
|
| rs2130748125 | 420 | E>V | No | Ensembl | |
| rs1269933229 | 422 | L>F | No |
TOPMed gnomAD |
|
| rs55785506 | 422 | L>P | No | Ensembl | |
| rs1210672549 | 426 | D>N | No |
TOPMed gnomAD |
|
| rs1210672549 | 426 | D>Y | No |
TOPMed gnomAD |
|
| rs760972252 | 428 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs750833038 | 428 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs779637244 | 429 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs779637244 | 429 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1810254598 | 430 | K>M | No | TOPMed | |
| rs1464396629 | 431 | K>Q | No | gnomAD | |
| rs1189221490 | 431 | K>R | No | gnomAD | |
| rs1263972733 | 432 | I>S | No | gnomAD | |
| rs1431633755 | 433 | P>A | No | gnomAD | |
| rs1810255080 | 433 | P>L | No | Ensembl | |
| rs1378572049 | 434 | P>A | No | gnomAD | |
| rs1810255398 | 435 | P>S | No | Ensembl | |
| rs754491610 | 437 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs1810255608 | 437 | N>K | No |
TOPMed gnomAD |
|
| rs34320206 | 439 | N>K | No |
ExAC gnomAD |
|
| rs748037623 | 440 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs2130767873 | 441 | A>T | No | Ensembl | |
| rs1810670274 | 442 | G>E | No | Ensembl | |
| rs1810670130 | 442 | G>R | No | gnomAD | |
| rs1447059041 | 443 | P>A | No | TOPMed | |
| rs754690758 | 444 | D>A | No |
ExAC gnomAD |
|
| rs754690758 | 444 | D>G | No |
ExAC gnomAD |
|
| rs1305929705 | 445 | D>G | No | gnomAD | |
| rs780516978 | 445 | D>N | No |
ExAC gnomAD |
|
| rs1305929705 | 445 | D>V | No | gnomAD | |
| rs150440508 | 446 | I>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150440508 | 446 | I>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1309134844 | 447 | R>K | No |
TOPMed gnomAD |
|
| rs368870320 | 448 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1810671617 | 448 | N>Y | No | Ensembl | |
| TCGA novel | 449 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1292636986 | 451 | T>I | No | gnomAD | |
| rs770672825 | 453 | F>L | No |
ExAC gnomAD |
|
| rs561540288 | 455 | E>D | No | Ensembl | |
| rs1251502985 | 458 | V>F | No | gnomAD | |
| rs1810672609 | 459 | P>A | No | gnomAD | |
| rs747155026 | 461 | S>C | No |
ExAC gnomAD |
|
| rs776788494 | 462 | V>L | No |
ExAC gnomAD |
|
| rs762048982 | 463 | C>Y | No |
ExAC gnomAD |
|
| rs1447930528 | 467 | D>N | No | TOPMed | |
| rs770408301 | 468 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1385371947 | 469 | S>A | No | gnomAD | |
| rs202151244 | 470 | I>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs763342872 | 470 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs202151244 | 470 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1442595003 | 473 | A>V | No | gnomAD | |
| rs1212943673 | 476 | L>M | No |
TOPMed gnomAD |
|
| TCGA novel | 476 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1810675184 | 476 | L>W | No | Ensembl | |
| rs1376841391 | 477 | E>Q | No | gnomAD | |
| rs1395411379 | 479 | D>G | No | gnomAD | |
| rs751975471 | 480 | D>Y | No |
ExAC gnomAD |
|
| rs375410364 | 481 | A>G | No |
ESP ExAC gnomAD |
|
| rs1810675672 | 482 | F>I | No | TOPMed | |
| rs369618696 | 483 | V>F | No |
ESP ExAC gnomAD |
|
| rs369618696 | 483 | V>I | No |
ESP ExAC gnomAD |
|
| rs777658022 | 488 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs777658022 | 488 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1437332474 | 490 | P>L | No |
TOPMed gnomAD |
|
| rs1273879443 | 490 | P>S | No | gnomAD | |
| rs1236116610 | 493 | D>H | No |
TOPMed gnomAD |
|
| rs1236116610 | 493 | D>N | No |
TOPMed gnomAD |
|
| rs753739226 | 496 | L>F | No |
ExAC gnomAD |
No associated diseases with Q96BR1
8 regional properties for Q96BR1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 162 - 419 | IPR000719 |
| domain | AGC-kinase, C-terminal | 420 - 496 | IPR000961 |
| domain | Phox homology | 12 - 124 | IPR001683 |
| active_site | Serine/threonine-protein kinase, active site | 282 - 294 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 168 - 191 | IPR017441 |
| domain | Protein kinase, C-terminal | 440 - 487 | IPR017892 |
| domain | Serine/threonine-protein kinase Sgk3, catalytic domain | 165 - 490 | IPR037709 |
| domain | CISK, PX domain | 12 - 120 | IPR037900 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.- | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early endosome | A membrane-bounded organelle that receives incoming material from primary endocytic vesicles that have been generated by clathrin-dependent and clathrin-independent endocytosis; vesicles fuse with the early endosome to deliver cargo for sorting into recycling or degradation pathways. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| recycling endosome | An organelle consisting of a network of tubules that functions in targeting molecules, such as receptors transporters and lipids, to the plasma membrane. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium channel regulator activity | Modulates the activity of a calcium channel. |
| chloride channel regulator activity | Binds to and modulates the activity of a chloride channel. |
| phosphatidylinositol binding | Binding to an inositol-containing glycerophospholipid, i.e. phosphatidylinositol (PtdIns) and its phosphorylated derivatives. |
| potassium channel regulator activity | Binds to and modulates the activity of a potassium channel. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
| sodium channel regulator activity | Binds to and modulates the activity of a sodium channel. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | Any process that stops, prevents or reduces the frequency, rate or extent of extrinsic apoptotic signaling pathway in absence of ligand. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of cell growth | Any process that modulates the frequency, rate, extent or direction of cell growth. |
| regulation of cell migration | Any process that modulates the frequency, rate or extent of cell migration. |
| regulation of cell population proliferation | Any process that modulates the frequency, rate or extent of cell proliferation. |
| regulation of DNA-binding transcription factor activity | Any process that modulates the frequency, rate or extent of the activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
42 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q6TJY3 | RPS6KB1 | Ribosomal protein S6 kinase beta-1 | Bos taurus (Bovine) | SS |
| P23443 | RPS6KB1 | Ribosomal protein S6 kinase beta-1 | Homo sapiens (Human) | EV SS |
| Q9UBS0 | RPS6KB2 | Ribosomal protein S6 kinase beta-2 | Homo sapiens (Human) | PR |
| Q96BR1 | SGK3 | Serine/threonine-protein kinase Sgk3 | Homo sapiens (Human) | SS |
| Q9Y5W7 | SNX14 | Sorting nexin-14 | Homo sapiens (Human) | PR |
| Q96S38 | RPS6KC1 | Ribosomal protein S6 kinase delta-1 | Homo sapiens (Human) | PR |
| Q9Y5W8 | SNX13 | Sorting nexin-13 | Homo sapiens (Human) | PR |
| O15530 | PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | Homo sapiens (Human) | EV |
| Q05513 | PRKCZ | Protein kinase C zeta type | Homo sapiens (Human) | SS |
| P41743 | PRKCI | Protein kinase C iota type | Homo sapiens (Human) | EV |
| Q16512 | PKN1 | Serine/threonine-protein kinase N1 | Homo sapiens (Human) | EV |
| Q6P5Z2 | PKN3 | Serine/threonine-protein kinase N3 | Homo sapiens (Human) | SS |
| Q16513 | PKN2 | Serine/threonine-protein kinase N2 | Homo sapiens (Human) | EV |
| P24723 | PRKCH | Protein kinase C eta type | Homo sapiens (Human) | SS |
| Q02156 | PRKCE | Protein kinase C epsilon type | Homo sapiens (Human) | SS |
| Q04759 | PRKCQ | Protein kinase C theta type | Homo sapiens (Human) | PR |
| Q05655 | PRKCD | Protein kinase C delta type | Homo sapiens (Human) | SS |
| P17252 | PRKCA | Protein kinase C alpha type | Homo sapiens (Human) | EV |
| P05129 | PRKCG | Protein kinase C gamma type | Homo sapiens (Human) | SS |
| P05771 | PRKCB | Protein kinase C beta type | Homo sapiens (Human) | SS |
| P31751 | AKT2 | RAC-beta serine/threonine-protein kinase | Homo sapiens (Human) | EV SS |
| P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| Q9Y243 | AKT3 | RAC-gamma serine/threonine-protein kinase | Homo sapiens (Human) | SS |
| Q9HBY8 | SGK2 | Serine/threonine-protein kinase Sgk2 | Homo sapiens (Human) | SS |
| O00141 | SGK1 | Serine/threonine-protein kinase Sgk1 | Homo sapiens (Human) | PR |
| Q15208 | STK38 | Serine/threonine-protein kinase 38 | Homo sapiens (Human) | EV |
| Q9Y2H1 | STK38L | Serine/threonine-protein kinase 38-like | Homo sapiens (Human) | EV |
| Q6A1A2 | PDPK2P | Putative 3-phosphoinositide-dependent protein kinase 2 | Homo sapiens (Human) | PR |
| Q8QZV4 | Stk32c | Serine/threonine-protein kinase 32C | Mus musculus (Mouse) | PR |
| Q9Z1M4 | Rps6kb2 | Ribosomal protein S6 kinase beta-2 | Mus musculus (Mouse) | SS |
| Q8BSK8 | Rps6kb1 | Ribosomal protein S6 kinase beta-1 | Mus musculus (Mouse) | SS |
| Q6PHS6 | Snx13 | Sorting nexin-13 | Mus musculus (Mouse) | PR |
| Q8BLK9 | Rps6kc1 | Ribosomal protein S6 kinase delta-1 | Mus musculus (Mouse) | PR |
| Q3ZT31 | Snx25 | Sorting nexin-25 | Mus musculus (Mouse) | PR |
| Q8BHY8 | Snx14 | Sorting nexin-14 | Mus musculus (Mouse) | PR |
| Q9ERE3 | Sgk3 | Serine/threonine-protein kinase Sgk3 | Mus musculus (Mouse) | PR |
| P67999 | Rps6kb1 | Ribosomal protein S6 kinase beta-1 | Rattus norvegicus (Rat) | EV SS |
| Q9SUA3 | D6PKL1 | Serine/threonine-protein kinase D6PKL1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O48963 | PHOT1 | Phototropin-1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64682 | PID | Protein kinase PINOID | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LUL2 | WAG2 | Serine/threonine-protein kinase WAG2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q5BKK4 | sgk1 | Serine/threonine-protein kinase Sgk1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MQRDHTMDYK | ESCPSVSIPS | SDEHREKKKR | FTVYKVLVSV | GRSEWFVFRR | YAEFDKLYNT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LKKQFPAMAL | KIPAKRIFGD | NFDPDFIKQR | RAGLNEFIQN | LVRYPELYNH | PDVRAFLQMD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SPKHQSDPSE | DEDERSSQKL | HSTSQNINLG | PSGNPHAKPT | DFDFLKVIGK | GSFGKVLLAK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RKLDGKFYAV | KVLQKKIVLN | RKEQKHIMAE | RNVLLKNVKH | PFLVGLHYSF | QTTEKLYFVL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DFVNGGELFF | HLQRERSFPE | HRARFYAAEI | ASALGYLHSI | KIVYRDLKPE | NILLDSVGHV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VLTDFGLCKE | GIAISDTTTT | FCGTPEYLAP | EVIRKQPYDN | TVDWWCLGAV | LYEMLYGLPP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FYCRDVAEMY | DNILHKPLSL | RPGVSLTAWS | ILEELLEKDR | QNRLGAKEDF | LEIQNHPFFE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SLSWADLVQK | KIPPPFNPNV | AGPDDIRNFD | TAFTEETVPY | SVCVSSDYSI | VNASVLEADD |
| 490 | |||||
| AFVGFSYAPP | SEDLFL |