Q96AX2
Gene name |
RAB37 |
Protein name |
Ras-related protein Rab-37 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:326624 |
EC number |
|
Protein Class |
|
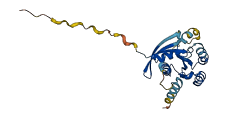
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q96AX2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q96AX2-F1 | Predicted | AlphaFoldDB |
188 variants for Q96AX2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs766791134 | 18 | R>G | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1385555512 CA400942476 |
39 | G>S | No |
ClinGen gnomAD |
|
|
CA400942489 rs750256354 |
40 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750192 rs750256354 |
40 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs149746610 CA8750194 |
41 | G>C | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs149746610 CA8750193 |
41 | G>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA400942522 rs746975143 |
44 | C>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750195 rs746975143 |
44 | C>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1248364080 CA400942602 |
51 | D>N | No |
ClinGen gnomAD |
|
|
rs1334325571 CA400942618 |
52 | G>E | No |
ClinGen TOPMed |
|
|
COSM1290755 CA8750197 rs777757102 CA293957990 |
52 | G>R | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs749330487 CA8750198 |
53 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs969539196 CA293957993 |
56 | S>F | No |
ClinGen TOPMed |
|
| TCGA novel | 57 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA8750201 rs746236671 CA400942659 |
57 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750203 rs772385837 |
58 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750202 rs772385837 |
58 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400942685 rs764620456 |
60 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400942687 rs1254894593 |
60 | I>T | No |
ClinGen gnomAD |
|
|
CA8750205 rs764620456 |
60 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400942704 rs1428644657 |
62 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
rs140394994 CA8750207 |
63 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs868407548 CA293958015 |
64 | G>D | No |
ClinGen Ensembl |
|
|
CA8750210 rs199750680 |
64 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750209 rs199750680 |
64 | G>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750222 rs772318569 |
70 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs560133058 CA8750223 |
71 | V>M | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA400932005 rs1358866411 |
72 | V>A | No |
ClinGen gnomAD |
|
| TCGA novel | 73 | T>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA400932068 rs1290711788 |
76 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
CA400932073 rs1290711788 |
76 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
rs145019795 CA8750228 |
77 | V>M | No |
ClinGen ESP ExAC gnomAD |
|
|
CA8750229 rs772822712 |
80 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750231 rs766017145 |
82 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
rs766017145 CA400932165 |
82 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1373729851 CA400932200 |
83 | I>T | No |
ClinGen gnomAD |
|
|
rs1225238831 CA400932203 |
84 | W>G | No |
ClinGen gnomAD |
|
|
CA293917663 rs910837302 |
84 | W>S | No |
ClinGen Ensembl |
|
|
CA8750246 rs772882123 COSM1479973 COSM1479972 COSM1479971 |
87 | A>T | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs146780541 CA8750247 |
88 | G>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8750249 rs770507152 |
91 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs759107202 CA8750250 |
91 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs770507152 COSM562159 CA8750248 COSM562161 COSM562160 |
91 | R>W | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM984111 COSM984113 CA8750252 rs150761775 COSM984112 |
93 | R>* | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs760647591 CA8750253 |
93 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1309701458 CA400932361 |
94 | S>G | No |
ClinGen gnomAD |
|
|
CA400932377 rs763968925 |
94 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM1385804 CA8750255 COSM1385805 COSM1385803 rs143690968 |
95 | V>I | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs929071371 CA293917720 |
97 | H>R | No |
ClinGen gnomAD |
|
|
rs1439304096 CA400932417 |
97 | H>Y | No |
ClinGen gnomAD |
|
|
COSM3796050 COSM3796048 COSM3796049 CA293917747 rs527644258 |
101 | R>K | Variant assessed as Somatic; 0.0 impact. urinary_tract [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes NCI-TCGA |
|
rs1403558405 CA400932512 |
102 | D>N | No |
ClinGen gnomAD |
|
|
rs1347003953 CA400932542 |
104 | Q>* | No |
ClinGen TOPMed |
|
|
CA293917813 rs71361632 |
106 | L>FV | No |
ClinGen Ensembl |
|
|
CA8750272 rs148142349 |
107 | L>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750274 rs761638080 |
108 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs909850875 CA400932583 |
109 | L>V | No |
ClinGen TOPMed |
|
|
rs1220928465 CA400932591 |
110 | Y>C | No |
ClinGen gnomAD |
|
|
CA400932597 rs1317747597 |
111 | D>N | No |
ClinGen gnomAD |
|
|
rs1436408440 CA400932622 |
114 | N>K | No |
ClinGen TOPMed |
|
|
CA8750277 rs755279933 |
116 | S>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs767801558 CA8750278 |
118 | F>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750279 rs767801558 |
118 | F>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750281 rs370945537 |
119 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1478781668 CA400932661 |
120 | N>I | No |
ClinGen gnomAD |
|
|
rs949231586 CA293917923 |
121 | I>S | No |
ClinGen TOPMed gnomAD |
|
|
CA8750282 rs749864575 |
122 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs757683419 CA8750283 |
122 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750284 rs757683419 |
122 | R>M | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 123 | A>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1303907212 CA400933574 |
124 | W>* | No |
ClinGen gnomAD |
|
|
rs751558943 CA293918646 |
125 | L>F | No |
ClinGen Ensembl |
|
|
rs200513132 CA400933586 |
126 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs200513132 CA8750307 |
126 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400933594 rs1232965464 |
127 | E>D | No |
ClinGen gnomAD |
|
|
rs1241391747 CA400933613 |
130 | E>V | No |
ClinGen TOPMed |
|
|
CA8750309 rs769786339 |
132 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA8750310 rs772174577 |
133 | Q>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400933640 rs1221144245 |
134 | R>K | No |
ClinGen TOPMed |
|
|
CA8750314 rs147358898 CA400933649 |
135 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750311 rs762982946 |
135 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA8750312 rs143298131 |
135 | D>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs762014090 CA8750317 |
136 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750315 rs764185728 |
136 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750316 rs764185728 |
136 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 136 | V>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA400933653 rs1182254894 |
137 | V>M | No |
ClinGen gnomAD |
|
|
rs1157534806 CA400933673 |
139 | M>I | No |
ClinGen gnomAD |
|
|
rs750982235 CA8750319 |
139 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750320 rs758877227 |
140 | L>R | No |
ClinGen ExAC gnomAD |
|
|
CA400933685 rs1295856023 |
142 | G>A | No |
ClinGen gnomAD |
|
|
CA8750321 rs779578722 |
142 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400933688 rs1479157700 |
143 | N>D | No |
ClinGen TOPMed |
|
|
CA293918775 rs112606240 |
143 | N>S | No |
ClinGen Ensembl |
|
|
rs1555595919 CA400933698 |
144 | K>T | No |
ClinGen Ensembl |
|
|
CA8750374 rs748208881 |
145 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400933717 rs748208881 |
145 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750375 rs770015617 |
146 | D>A | No |
ClinGen ExAC gnomAD |
|
|
rs773354446 CA8750376 |
147 | M>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400933727 rs1358558768 |
147 | M>K | No |
ClinGen gnomAD |
|
|
CA293919239 rs936156108 |
147 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1233402952 CA400933735 |
148 | S>N | No |
ClinGen TOPMed |
|
|
rs1369400532 CA400933742 |
149 | S>G | No |
ClinGen gnomAD |
|
|
rs201839620 CA293919245 |
149 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 150 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs774756333 CA8750379 |
150 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs137872401 CA8750378 |
150 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400933757 rs1241703012 |
151 | R>K | No |
ClinGen gnomAD |
|
| TCGA novel | 151 | R>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA400933764 rs1598331248 |
152 | V>A | No |
ClinGen Ensembl |
|
|
CA400933765 rs1598331248 |
152 | V>G | No |
ClinGen Ensembl |
|
|
rs907993927 CA293919287 |
153 | I>T | No |
ClinGen Ensembl |
|
|
CA293919294 rs543387119 |
154 | R>C | No |
ClinGen 1000Genomes gnomAD |
|
|
rs149486886 CA8750380 |
154 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs149486886 CA8750381 |
154 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs149486886 CA400933774 |
154 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8750384 rs774889397 |
156 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA400933792 rs895360538 |
157 | D>E | No |
ClinGen TOPMed |
|
|
CA8750385 rs760208960 |
157 | D>H | No |
ClinGen ExAC gnomAD |
|
|
CA8750386 rs760208960 |
157 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs143915644 CA8750388 |
158 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400933799 rs1424800509 |
159 | E>Q | No |
ClinGen gnomAD |
|
|
CA400933809 rs1567817135 |
160 | T>A | No |
ClinGen Ensembl |
|
|
CA400933817 rs756945292 |
160 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA8750389 rs756945292 |
160 | T>N | No |
ClinGen ExAC gnomAD |
|
|
rs1160533812 CA400933829 |
161 | L>F | No |
ClinGen gnomAD |
|
|
rs1427279841 CA400933826 |
161 | L>S | No |
ClinGen TOPMed |
|
|
rs374935761 CA8750390 |
163 | R>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400933850 rs1598331346 |
163 | R>S | No |
ClinGen Ensembl |
|
|
rs963741004 CA293919502 |
164 | E>G | No |
ClinGen Ensembl |
|
|
rs762740730 CA400933916 |
165 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs867840244 CA293919509 |
166 | G>D | No |
ClinGen Ensembl |
|
|
rs766081840 CA8750411 |
166 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751557494 CA8750412 |
168 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA400933993 rs1328925117 |
172 | T>N | No |
ClinGen gnomAD |
|
|
CA400934010 rs977071609 |
173 | S>R | No |
ClinGen gnomAD |
|
|
CA400933999 rs1375693483 |
173 | S>R | No |
ClinGen gnomAD |
|
|
CA8750413 rs754889461 |
174 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1227060514 CA400934043 |
176 | T>A | No |
ClinGen gnomAD |
|
|
rs777871045 CA8750414 |
176 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750415 rs148637613 |
178 | M>V | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1167229844 CA400934154 |
181 | E>D | No |
ClinGen TOPMed |
|
|
rs779143134 CA8750417 |
182 | L>* | No |
ClinGen ExAC gnomAD |
|
|
CA400934182 rs1256474430 |
182 | L>F | No |
ClinGen gnomAD |
|
|
rs1567817307 CA400934248 |
186 | A>T | No |
ClinGen Ensembl |
|
|
rs772557537 CA8750419 |
187 | I>M | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 187 | I>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs746225239 CA8750418 |
187 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs34215331 VAR_034434 CA293919631 |
188 | A>P | No |
ClinGen UniProt ExAC TOPMed dbSNP gnomAD |
|
|
rs34215331 CA8750421 |
188 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775475701 CA8750462 |
190 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs775475701 CA400934408 |
190 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1452003879 CA400934431 |
191 | L>R | No |
ClinGen gnomAD |
|
|
CA293919960 rs914677226 |
193 | Y>N | No |
ClinGen TOPMed |
|
|
CA8750466 rs150021744 |
194 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8750465 rs150021744 |
194 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8750464 rs765394498 |
194 | R>W | No |
ClinGen ExAC TOPMed |
|
|
CA8750468 CA8750469 rs139440620 |
196 | G>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM361180 CA400934509 COSM361181 COSM361182 rs1158729238 |
196 | G>V | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs781678010 CA400934514 |
197 | H>D | No |
ClinGen ExAC gnomAD |
|
|
rs1567817566 CA400934523 |
197 | H>Q | No |
ClinGen Ensembl |
|
|
CA8750471 rs200335281 |
197 | H>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs781678010 CA8750470 |
197 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA8750472 rs756549615 |
198 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs368389878 CA8750473 |
198 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400934551 rs748698308 |
199 | A>E | No |
ClinGen ExAC gnomAD |
|
|
rs748698308 CA8750474 |
199 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs534120632 CA8750476 |
200 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750477 rs745519873 |
201 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1310103092 CA400934606 |
202 | P>L | No |
ClinGen gnomAD |
|
|
COSM1385811 COSM1385809 COSM1385810 CA400934658 rs1356004989 |
205 | Q>R | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA8750478 rs771915095 |
206 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs760695554 CA400934690 |
207 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763884748 CA8750481 |
207 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs202002183 CA8750482 |
208 | D>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA293920073 rs951347274 |
209 | Y>D | No |
ClinGen TOPMed gnomAD |
|
|
CA293920065 rs951347274 |
209 | Y>H | No |
ClinGen TOPMed gnomAD |
|
|
CA8750484 rs766562014 |
211 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400934783 rs1180452795 |
212 | S>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs898251293 CA293920079 |
214 | K>E | No |
ClinGen TOPMed |
|
|
rs759808920 CA8750486 |
215 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
rs768035222 CA8750487 |
216 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs537134838 CA8750488 |
216 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1175113339 CA400934859 |
220 | C>G | No |
ClinGen TOPMed |
|
|
rs201555817 CA8750489 |
221 | S>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 222 | F>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1306312077 CA400934925 |
223 | M>I | No |
ClinGen gnomAD |
|
|
CA8750490 rs778084858 |
223 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
No associated diseases with Q96AX2
1 regional properties for Q96AX2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 29 - 184 | IPR005225 |
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| azurophil granule membrane | The lipid bilayer surrounding an azurophil granule, a primary lysosomal granule found in neutrophil granulocytes that contains a wide range of hydrolytic enzymes and is released into the extracellular fluid. |
| endoplasmic reticulum-Golgi intermediate compartment | A complex system of membrane-bounded compartments located between endoplasmic reticulum (ER) and the Golgi complex, with a distinctive membrane protein composition; involved in ER-to-Golgi and Golgi-to-ER transport. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| specific granule membrane | The lipid bilayer surrounding a specific granule, a granule with a membranous, tubular internal structure, found primarily in mature neutrophil cells. Most are released into the extracellular fluid. Specific granules contain lactoferrin, lysozyme, vitamin B12 binding protein and elastase. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
9 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9H082 | RAB33B | Ras-related protein Rab-33B | Homo sapiens (Human) | PR |
| Q9JKM7 | Rab37 | Ras-related protein Rab-37 | Mus musculus (Mouse) | PR |
| P25766 | RGP1 | Ras-related protein RGP1 | Oryza sativa subsp japonica (Rice) | PR |
| Q9LNK1 | RABA3 | Ras-related protein RABA3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O04486 | RABA2A | Ras-related protein RABA2a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LNW1 | RABA2B | Ras-related protein RABA2b | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LH50 | RABA4D | Ras-related protein RABA4d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FE79 | RABA4C | Ras-related protein RABA4c | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJZ5 | RABA4E | Putative Ras-related protein RABA4e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTGTPGAVAT | RDGEAPERSP | PCSPSYDLTG | KVMLLGDTGV | GKTCFLIQFK | DGAFLSGTFI |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ATVGIDFRNK | VVTVDGVRVK | LQIWDTAGQE | RFRSVTHAYY | RDAQALLLLY | DITNKSSFDN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IRAWLTEIHE | YAQRDVVIML | LGNKADMSSE | RVIRSEDGET | LAREYGVPFL | ETSAKTGMNV |
| 190 | 200 | 210 | 220 | ||
| ELAFLAIAKE | LKYRAGHQAD | EPSFQIRDYV | ESQKKRSSCC | SFM |