Q969Q4
Gene name |
ARL11 (ARLTS1) |
Protein name |
ADP-ribosylation factor-like protein 11 |
Names |
ADP-ribosylation factor-like tumor suppressor protein 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:115761 |
EC number |
|
Protein Class |
|
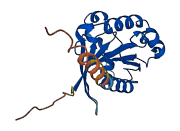
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q969Q4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q969Q4-F1 | Predicted | AlphaFoldDB |
195 variants for Q969Q4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs201791650 CA6983068 |
4 | V>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983067 rs772364057 |
4 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1369947113 CA388192340 |
6 | S>F | No |
ClinGen gnomAD |
|
|
CA388192407 rs1271815383 |
9 | H>P | No |
ClinGen gnomAD |
|
|
rs12584992 CA249369357 |
10 | K>N | No |
ClinGen gnomAD |
|
|
COSM3417647 rs566163670 CA6983069 |
11 | A>V | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
CA388192555 rs1469469563 |
14 | Q>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA6983071 rs751603349 |
14 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs370668058 CA6983073 |
16 | V>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA388192726 rs1475383091 |
18 | M>I | No |
ClinGen gnomAD |
|
|
rs758066262 CA6983075 |
18 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388192761 rs1166167152 |
19 | G>V | No |
ClinGen gnomAD |
|
|
CA388192805 rs1462086461 |
21 | D>A | No |
ClinGen gnomAD |
|
|
CA6983076 rs777360422 |
21 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA6983078 rs147389782 |
22 | S>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983077 rs147389782 VAR_023742 |
22 | S>L | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs769707452 CA388192866 |
23 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6983081 rs769707452 COSM432471 |
23 | A>V | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs536056437 CA6983083 |
24 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs771958819 CA249369383 |
26 | T>P | No |
ClinGen Ensembl |
|
|
COSM190487 CA388192936 rs1394373519 |
27 | T>M | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
CA388192951 rs1311937193 |
28 | L>P | No |
ClinGen gnomAD |
|
|
rs576032054 CA249369390 |
30 | Y>C | No |
ClinGen 1000Genomes |
|
|
CA6983087 rs201132357 |
32 | L>P | No |
ClinGen 1000Genomes ExAC |
|
| TCGA novel | 34 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs776703350 CA6983088 |
34 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA388193018 rs1414929363 |
38 | V>A | No |
ClinGen TOPMed |
|
|
CA6983089 rs759972408 |
39 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201969290 CA6983090 |
40 | T>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1468766993 CA388193027 |
40 | T>P | No |
ClinGen TOPMed |
|
|
CA6983092 rs763346893 |
42 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1420702453 CA388193052 |
44 | V>A | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 45 | G>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA388193055 rs1338502851 |
45 | G>R | No |
ClinGen TOPMed |
|
|
CA388193054 rs1338502851 |
45 | G>S | No |
ClinGen TOPMed |
|
|
CA388193074 rs751269133 |
47 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1566301710 CA388193077 |
48 | V>E | No |
ClinGen Ensembl |
|
|
rs756928549 CA6983096 |
48 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6983095 rs756928549 |
48 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1415880073 CA388193084 |
49 | E>G | No |
ClinGen gnomAD |
|
|
rs1290304131 CA388193082 |
49 | E>K | No |
ClinGen TOPMed |
|
|
rs1566301716 CA388193096 |
51 | L>R | No |
ClinGen Ensembl |
|
|
rs996300560 CA249369409 |
53 | A>T | No |
ClinGen TOPMed |
|
|
CA388193121 rs1455540602 |
54 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA6983097 rs749892930 |
57 | V>M | No |
ClinGen ExAC gnomAD |
|
|
COSM947898 CA249369417 rs958511445 |
61 | L>I | Variant assessed as Somatic; impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated Ensembl NCI-TCGA |
|
rs1357327382 CA388193288 |
64 | V>A | No |
ClinGen gnomAD |
|
|
CA6983099 rs143660006 COSM1367379 |
64 | V>I | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
CA6983101 rs117251022 |
65 | G>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA388193301 rs117251022 |
65 | G>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983100 rs117251022 |
65 | G>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA388193314 rs1332128095 |
66 | G>E | No |
ClinGen gnomAD |
|
|
CA6983102 rs778840264 |
66 | G>R | No |
ClinGen ExAC gnomAD |
|
| rs1324509298 | 67 | Q>A | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA388193334 rs1566301752 |
67 | Q>L | No |
ClinGen Ensembl |
|
|
CA6983103 rs747265258 |
68 | A>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs113027020 CA6983105 |
69 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771123171 CA6983104 |
69 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388193392 rs1187226142 |
71 | R>K | No |
ClinGen gnomAD |
|
|
rs1255483525 CA388193402 |
71 | R>S | No |
ClinGen gnomAD |
|
|
CA388193426 rs1566301777 |
73 | S>N | No |
ClinGen Ensembl |
|
|
rs776025417 CA6983109 |
74 | W>L | No |
ClinGen ExAC gnomAD |
|
|
rs763436324 CA6983110 |
77 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA388193527 rs1156640192 |
78 | L>R | No |
ClinGen gnomAD |
|
|
rs149745507 CA6983111 |
78 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs761386601 CA6983113 |
79 | E>* | No |
ClinGen ExAC gnomAD |
|
|
CA388193541 rs1435268015 |
79 | E>G | No |
ClinGen gnomAD |
|
|
rs767150266 CA6983114 |
80 | G>S | No |
ClinGen ExAC |
|
| TCGA novel | 82 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA388193572 rs1392676444 |
83 | I>S | No |
ClinGen TOPMed gnomAD |
|
|
CA388193590 rs1360753264 |
84 | L>P | No |
ClinGen gnomAD |
|
|
CA388193606 rs1237259832 |
85 | V>A | No |
ClinGen gnomAD |
|
|
rs374531869 CA6983117 |
85 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM1367382 CA6983116 rs374531869 |
85 | V>M | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA388193630 rs377295532 |
86 | Y>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs370030859 CA6983119 |
87 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA249369444 COSM1514074 rs867820824 |
89 | D>N | lung [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs200663291 CA6983120 |
91 | T>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA6983121 rs747926100 |
91 | T>I | No |
ClinGen ExAC TOPMed |
|
|
rs757447757 CA6983122 |
92 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 92 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 93 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs781514079 CA6983123 |
94 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs544417615 CA6983124 |
95 | R>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA6983125 rs769920049 |
95 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1176478777 CA388193769 |
96 | L>S | No |
ClinGen gnomAD |
|
|
CA6983128 rs769095684 |
98 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs774603360 CA6983129 |
99 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772293845 CA388193836 |
100 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772293845 CA6983131 |
100 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388193906 rs1216934556 |
103 | L>P | No |
ClinGen TOPMed |
|
|
CA388193911 rs1485136473 |
104 | T>P | No |
ClinGen TOPMed |
|
|
CA6983136 rs759566544 |
109 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753334860 CA6983135 |
109 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs532945151 CA6983137 |
110 | P>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA6983138 rs752542486 |
111 | N>D | No |
ClinGen ExAC gnomAD |
|
|
rs1301834194 CA388194041 |
111 | N>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs761431152 CA6983139 |
112 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777416619 CA388194066 |
113 | A>P | No |
ClinGen ExAC gnomAD |
|
|
rs777416619 CA6983140 |
113 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA6983141 rs750723672 |
113 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA388194077 rs1479737976 |
114 | G>S | No |
ClinGen gnomAD |
|
|
CA388194098 rs1253922375 |
115 | V>D | No |
ClinGen gnomAD |
|
|
CA388194101 rs1253922375 |
115 | V>G | No |
ClinGen gnomAD |
|
|
CA249369481 rs994780789 |
115 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs780120181 CA6983143 |
119 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA249369488 VAR_048318 rs35712316 |
120 | L>M | No |
ClinGen UniProt Ensembl dbSNP |
|
|
CA388194180 rs868285879 |
121 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA249369492 rs868285879 |
121 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1171133622 CA388194201 |
122 | N>K | No |
ClinGen gnomAD |
|
|
rs1031176774 CA249369494 |
125 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
CA388194255 rs1463649160 |
125 | E>G | No |
ClinGen gnomAD |
|
|
rs1371983031 CA388194247 |
125 | E>Q | No |
ClinGen gnomAD |
|
|
CA6983145 rs769184018 |
126 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs866872755 CA249369498 |
127 | P>L | No |
ClinGen Ensembl |
|
|
CA6983146 rs779514414 |
129 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6983147 rs748581908 |
129 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA388194312 rs1566301940 |
130 | L>F | No |
ClinGen Ensembl |
|
|
VAR_023743 rs147120792 CA6983148 |
131 | P>L | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA388194330 rs147120792 |
131 | P>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs770655477 CA6983151 |
133 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA388194365 rs1235262427 |
134 | K>E | No |
ClinGen gnomAD |
|
|
CA388194363 rs1235262427 |
134 | K>Q | No |
ClinGen gnomAD |
|
|
CA6983152 rs776309613 |
134 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA388194393 rs1334456532 |
135 | I>M | No |
ClinGen gnomAD |
|
|
CA6983153 rs759152923 |
136 | R>T | No |
ClinGen ExAC gnomAD |
|
|
rs970805802 CA249369500 |
137 | N>H | No |
ClinGen Ensembl |
|
|
rs764627150 CA6983154 |
137 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752737224 CA6983155 |
138 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388194432 rs1187009656 |
138 | R>K | No |
ClinGen gnomAD |
|
|
CA249369501 rs766738083 |
139 | L>P | No |
ClinGen Ensembl |
|
|
CA388194462 rs1594391878 |
140 | S>N | No |
ClinGen Ensembl |
|
|
CA249369502 rs923965064 |
142 | E>G | No |
ClinGen gnomAD |
|
|
rs762794413 CA6983156 |
142 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA6983157 rs763952974 |
144 | F>Y | No |
ClinGen ExAC gnomAD |
|
|
CA6983158 rs751295835 |
145 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA388194604 rs567358654 |
148 | C>* | No |
ClinGen 1000Genomes gnomAD |
|
|
CA388194594 rs3803185 |
148 | C>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983160 VAR_023744 rs3803185 |
148 | C>R | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs567358654 CA249369507 |
148 | C>W | No |
ClinGen 1000Genomes gnomAD |
|
|
RCV000948924 CA6983161 COSM4147728 rs34301344 |
149 | W>* | thyroid [Cosmic] | No |
ClinGen cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs755100942 CA6983162 COSM4147728 |
149 | W>* | thyroid [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
| TCGA novel | 150 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs867095637 CA249369509 |
150 | E>K | No |
ClinGen Ensembl |
|
|
rs1339136847 CA388194654 |
151 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
CA249369511 rs915492243 |
152 | R>Q | No |
ClinGen gnomAD |
|
|
rs778849305 CA6983163 COSM947899 |
152 | R>W | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA6983164 rs748760701 |
153 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758766792 CA6983165 |
154 | C>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758766792 CA388194683 |
154 | C>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388194710 rs1286667760 |
155 | S>R | No |
ClinGen gnomAD |
|
|
CA6983166 rs778216279 |
156 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6983167 rs747335286 |
157 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA388194748 rs1396778377 |
158 | T>N | No |
ClinGen TOPMed |
|
|
CA388194765 rs1395927294 |
160 | E>A | No |
ClinGen TOPMed |
|
|
rs551214371 CA6983169 |
160 | E>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 163 | P>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA6983170 rs745498908 |
163 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1171214636 CA388194823 |
164 | E>A | No |
ClinGen TOPMed |
|
|
CA6983172 COSM1238229 VAR_023745 rs146850453 |
164 | E>K | oesophagus [Cosmic] | No |
ClinGen cosmic curated UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
CA388194813 rs146850453 |
164 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs911644636 CA249369523 |
165 | A>D | No |
ClinGen TOPMed |
|
|
rs911644636 CA249369521 |
165 | A>G | No |
ClinGen TOPMed |
|
|
CA6983173 rs762961641 |
167 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA388194950 rs1409762273 |
167 | Q>P | No |
ClinGen gnomAD |
|
|
CA388195023 rs1158867167 |
169 | L>R | No |
ClinGen gnomAD |
|
|
rs774163543 CA6983175 |
170 | W>R | No |
ClinGen ExAC gnomAD |
|
|
CA388195154 rs1354877624 |
173 | L>Q | No |
ClinGen gnomAD |
|
|
rs767279791 CA6983177 |
176 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6983179 rs140526352 |
176 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983178 rs140526352 |
176 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA388195325 rs1594392012 |
178 | C>Y | No |
ClinGen Ensembl |
|
|
rs1566302128 CA388195367 |
179 | M>I | No |
ClinGen Ensembl |
|
|
CA6983181 rs752665403 |
179 | M>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752665403 CA388195362 |
179 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765365695 CA6983180 |
179 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs758972109 COSM1188716 CA6983182 |
182 | Q>* | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA6983183 rs115070474 |
183 | A>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983184 rs115070474 |
183 | A>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983186 rs781685676 |
184 | R>T | No |
ClinGen ExAC gnomAD |
|
|
CA6983187 rs745519193 |
185 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs114223009 CA6983190 |
186 | H>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983189 rs115584350 |
186 | H>Y | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6983192 rs369303916 |
190 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA6983193 rs574095059 |
190 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs562619284 CA6983195 |
191 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs200112422 CA6983196 |
192 | D>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs577463512 CA249369559 |
192 | D>H | No |
ClinGen 1000Genomes |
|
|
rs765332310 CA388195761 |
193 | S>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765332310 CA6983197 |
193 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1480613829 CA388195764 |
193 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA388195759 rs765332310 |
193 | S>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6983198 rs35835937 |
194 | K>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1441750086 CA388195777 |
194 | K>R | No |
ClinGen gnomAD |
|
|
CA388195796 rs1458149068 |
195 | R>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
No associated diseases with Q969Q4
8 regional properties for Q969Q4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | C2 domain | 666 - 794 | IPR000008 |
| domain | Phosphatidylinositol-specific phospholipase C, X domain | 312 - 464 | IPR000909 |
| domain | Phospholipase C, phosphatidylinositol-specific, Y domain | 550 - 666 | IPR001711 |
| domain | Phospholipase C-beta, C-terminal domain | 977 - 1151 | IPR014815 |
| domain | Phosphoinositide-specific phospholipase C, EF-hand-like domain | 214 - 303 | IPR015359 |
| domain | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2, catalytic domain | 311 - 653 | IPR028403 |
| domain | PLC-beta, PH domain | 12 - 144 | IPR037862 |
| domain | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2, EF-hand domain | 149 - 299 | IPR046969 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| hematopoietic progenitor cell differentiation | The process in which precursor cell type acquires the specialized features of a hematopoietic progenitor cell, a class of cell types including myeloid progenitor cells and lymphoid progenitor cells. |
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
48 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJ96 | ARL5A | ADP-ribosylation factor-like protein 5A | Bos taurus (Bovine) | SS |
| P84081 | ARF2 | ADP-ribosylation factor 2 | Bos taurus (Bovine) | SS |
| P84080 | ARF1 | ADP-ribosylation factor 1 | Bos taurus (Bovine) | SS |
| Q3SZF2 | ARF4 | ADP-ribosylation factor 4 | Bos taurus (Bovine) | SS |
| Q0VC18 | ARL4D | ADP-ribosylation factor-like protein 4D | Bos taurus (Bovine) | PR |
| P49702 | ARF5 | ADP-ribosylation factor 5 | Gallus gallus (Chicken) | SS |
| P40945 | Arf4 | ADP ribosylation factor 4 | Drosophila melanogaster (Fruit fly) | SS |
| P61209 | Arf1 | ADP-ribosylation factor 1 | Drosophila melanogaster (Fruit fly) | SS |
| A6NH57 | ARL5C | Putative ADP-ribosylation factor-like protein 5C | Homo sapiens (Human) | SS |
| P18085 | ARF4 | ADP-ribosylation factor 4 | Homo sapiens (Human) | EV |
| P49703 | ARL4D | ADP-ribosylation factor-like protein 4D | Homo sapiens (Human) | PR |
| P84077 | ARF1 | ADP-ribosylation factor 1 | Homo sapiens (Human) | EV |
| P84085 | ARF5 | ADP-ribosylation factor 5 | Homo sapiens (Human) | EV |
| Q8N4G2 | ARL14 | ADP-ribosylation factor-like protein 14 | Homo sapiens (Human) | SS |
| Q96KC2 | ARL5B | ADP-ribosylation factor-like protein 5B | Homo sapiens (Human) | SS |
| P62330 | ARF6 | ADP-ribosylation factor 6 | Homo sapiens (Human) | EV |
| Q8IVW1 | ARL17A | ADP-ribosylation factor-like protein 17 | Homo sapiens (Human) | SS |
| Q9H0F7 | ARL6 | ADP-ribosylation factor-like protein 6 | Homo sapiens (Human) | SS |
| P40616 | ARL1 | ADP-ribosylation factor-like protein 1 | Homo sapiens (Human) | SS |
| P61204 | ARF3 | ADP-ribosylation factor 3 | Homo sapiens (Human) | EV |
| Q9Y689 | ARL5A | ADP-ribosylation factor-like protein 5A | Homo sapiens (Human) | SS |
| P49076 | ARF1 | ADP-ribosylation factor | Zea mays (Maize) | SS |
| P84084 | Arf5 | ADP-ribosylation factor 5 | Mus musculus (Mouse) | SS |
| P61750 | Arf4 | ADP-ribosylation factor 4 | Mus musculus (Mouse) | SS |
| Q99PE9 | Arl4d | ADP-ribosylation factor-like protein 4D | Mus musculus (Mouse) | PR |
| Q6P068 | Arl5c | ADP-ribosylation factor-like protein 5C | Mus musculus (Mouse) | SS |
| Q80ZU0 | Arl5a | ADP-ribosylation factor-like protein 5A | Mus musculus (Mouse) | SS |
| P61205 | Arf3 | ADP-ribosylation factor 3 | Mus musculus (Mouse) | SS |
| Q9D4P0 | Arl5b | ADP-ribosylation factor-like protein 5B | Mus musculus (Mouse) | SS |
| P84078 | Arf1 | ADP-ribosylation factor 1 | Mus musculus (Mouse) | SS |
| Q8BSL7 | Arf2 | ADP-ribosylation factor 2 | Mus musculus (Mouse) | SS |
| P51824 | ADP-ribosylation factor 1 | Solanum tuberosum (Potato) | SS | |
| P84082 | Arf2 | ADP-ribosylation factor 2 | Rattus norvegicus (Rat) | SS |
| P51646 | Arl5a | ADP-ribosylation factor-like protein 5A | Rattus norvegicus (Rat) | SS |
| P84083 | Arf5 | ADP-ribosylation factor 5 | Rattus norvegicus (Rat) | SS |
| P36407 | Trim23 | E3 ubiquitin-protein ligase TRIM23 | Rattus norvegicus (Rat) | SS |
| P61751 | Arf4 | ADP-ribosylation factor 4 | Rattus norvegicus (Rat) | SS |
| P61206 | Arf3 | ADP-ribosylation factor 3 | Rattus norvegicus (Rat) | SS |
| P84079 | Arf1 | ADP-ribosylation factor 1 | Rattus norvegicus (Rat) | SS |
| P51823 | ARF | ADP-ribosylation factor 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q06396 | Os01g0813400 | ADP-ribosylation factor 1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q10943 | arf-1.2 | ADP-ribosylation factor 1-like 2 | Caenorhabditis elegans | SS |
| P34212 | arl-5 | ADP-ribosylation factor-like protein 5 | Caenorhabditis elegans | SS |
| P40940 | ARF3 | ADP-ribosylation factor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P36397 | ARF1 | ADP-ribosylation factor 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P0DH91 | ARF2-B | ADP-ribosylation factor 2-B | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LQC8 | ARF2-A | ADP-ribosylation factor 2-A | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5M9P8 | arl6 | ADP-ribosylation factor-like protein 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGSVNSRGHK | AEAQVVMMGL | DSAGKTTLLY | KLKGHQLVET | LPTVGFNVEP | LKAPGHVSLT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LWDVGGQAPL | RASWKDYLEG | TDILVYVLDS | TDEARLPESA | AELTEVLNDP | NMAGVPFLVL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ANKQEAPDAL | PLLKIRNRLS | LERFQDHCWE | LRGCSALTGE | GLPEALQSLW | SLLKSRSCMC |
| 190 | |||||
| LQARAHGAER | GDSKRS |