Q96576
Gene name |
PME3 |
Protein name |
Pectinesterase 3 |
Names |
PE 3, Pectin methylesterase 3 |
Species |
Solanum lycopersicum (Tomato) (Lycopersicon esculentum) |
KEGG Pathway |
|
EC number |
3.1.1.11: Carboxylic ester hydrolases |
Protein Class |
|
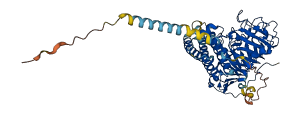
Descriptions
(Annotation from UniProt)
The PMEI region may act as an autoinhibitory domain.
Autoinhibitory domains (AIDs)
Target domain |
233-527 (Pectinesterase domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for Q96576
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q96576-F1 | Predicted | AlphaFoldDB |
No variants for Q96576
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q96576 | |||||
No associated diseases with Q96576
4 regional properties for Q96576
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.1.11 | Carboxylic ester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| aspartyl esterase activity | Catalysis of the hydrolysis of an ester bond by a mechanism involving a catalytically active aspartic acid residue. |
| pectinesterase activity | Catalysis of the reaction: pectin + n H2O = n methanol + pectate. |
| pectinesterase inhibitor activity | Binds to and stops, prevents or reduces the activity of pectinesterase. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| cell wall modification | The series of events leading to chemical and structural alterations of an existing cell wall that can result in loosening, increased extensibility or disassembly. |
| fruit ripening | An developmental maturation process that has as participant a fruit. Ripening causes changes in one or more characteristics of a fruit (color, aroma, flavor, texture, hardness, cell wall structure) and may make it more attractive to animals and aid in seed dispersal. |
| pectin catabolic process | The chemical reactions and pathways resulting in the breakdown of pectin, a polymer containing a backbone of alpha-1,4-linked D-galacturonic acid residues. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q1JPL7 | PME18 | Pectinesterase/pectinesterase inhibitor 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1PEC0 | PME42 | Probable pectinesterase/pectinesterase inhibitor 42 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84JX1 | PME19 | Probable pectinesterase/pectinesterase inhibitor 19 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P09607 | PME2.1 | Pectinesterase 2.1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| P14280 | PME1.9 | Pectinesterase 1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q43143 | PMEU1 | Pectinesterase/pectinesterase inhibitor U1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q96575 | PME2.2 | Pectinesterase 2.2 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MATPLQPFLT | KTHKQNPIIG | FNILTFVVTL | FVALFLVVFL | VAPYQFEIKH | SNLCKTAQDS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QLCLSYVSEI | VTTESDGVTV | LKKFLVKYVH | QMNNAIPVVR | KIKNQINDIR | QQGALTDCLE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LLDQSVDLVS | DSIAAIDKRS | RSEHANAQSW | LSGVLTNHVT | CLDELTSFSL | STKNGTVLDE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LITRAKVALA | MLASVTTPND | EVLRQGLGKM | PYWVSSRDRK | LMESSGKDII | ANRVVAQDGT |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GDYQTLAEAV | AAAPDKNKTR | YVIYVKMGIY | KENVVVTKKK | MNLMIVGDGM | NATIITGSLN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VVDGSTFPSN | TLAAVGQGFI | LQDICIQNTA | GPEKDQAVAL | RVGADMSVIN | RCRIDAYQDT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LYAHSQRQFY | RDSYVTGTVD | FIFGNAAVVF | QKCQIVARKP | NKRQKNMVTA | QGRTDPNQAT |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GTSIQFCDII | ASPDLEPVMN | EYKTYLGRPW | KKHSRTVVMQ | SYLDGHIDPS | GWFEWRGDFA |
| 490 | 500 | 510 | 520 | 530 | 540 |
| LKTLYYGEFM | NNGPGAGTSK | RVKWPGYHVI | TDPNEAMPFT | VAELIQGGSW | LNSTSVAYVE |
| GLVE |