Q94C77
Gene name |
At4g34220 (F10M10.12, F28A23_20) |
Protein name |
Receptor protein kinase-like protein At4g34220 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT4G34220 |
EC number |
|
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
622-646 (Activation loop from InterPro)
Target domain |
471-753 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
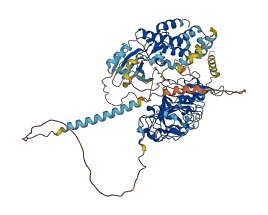
Autoinhibited structure

Activated structure

1 structures for Q94C77
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q94C77-F1 | Predicted | AlphaFoldDB |
63 variants for Q94C77
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
ENSVATH12384671 ENSVATH00551869 |
6 | S>R | No | 1000Genomes | |
| ENSVATH12384670 | 33 | L>I | No | 1000Genomes | |
| tmp_4_16383941_T_G | 38 | K>N | No | 1000Genomes | |
| tmp_4_16383927_G_C | 43 | T>S | No | 1000Genomes | |
| tmp_4_16383875_G_T | 60 | C>* | No | 1000Genomes | |
| ENSVATH06822357 | 71 | G>A | No | 1000Genomes | |
| tmp_4_16383828_G_C | 76 | P>R | No | 1000Genomes | |
| ENSVATH06822356 | 105 | L>I | No | 1000Genomes | |
| ENSVATH06822355 | 141 | G>A | No | 1000Genomes | |
| ENSVATH02956547 | 143 | L>M | No | 1000Genomes | |
| ENSVATH00551865 | 149 | S>R | No | 1000Genomes | |
| ENSVATH00551864 | 166 | E>Q | No | 1000Genomes | |
| ENSVATH06822353 | 183 | S>L | No | 1000Genomes | |
| ENSVATH06822354 | 183 | S>T | No | 1000Genomes | |
| ENSVATH02956544 | 186 | T>S | No | 1000Genomes | |
| ENSVATH02956543 | 201 | L>F | No | 1000Genomes | |
| tmp_4_16383436_G_T | 207 | L>I | No | 1000Genomes | |
| tmp_4_16383427_C_T | 210 | G>S | No | 1000Genomes | |
| ENSVATH00551863 | 238 | N>D | No | 1000Genomes | |
| ENSVATH02956542 | 238 | N>S | No | 1000Genomes | |
| ENSVATH02956541 | 262 | S>N | No | 1000Genomes | |
| ENSVATH12384668 | 267 | L>F | No | 1000Genomes | |
| ENSVATH02956540 | 280 | L>F | No | 1000Genomes | |
| ENSVATH02956538 | 323 | T>S | No | 1000Genomes | |
| ENSVATH00551858 | 325 | K>T | No | 1000Genomes | |
| tmp_4_16383021_G_A | 345 | A>V | No | 1000Genomes | |
| ENSVATH00551857 | 348 | V>A | No | 1000Genomes | |
| tmp_4_16382995_C_A | 354 | G>C | No | 1000Genomes | |
| ENSVATH02956537 | 381 | C>Y | No | 1000Genomes | |
| tmp_4_16382886_G_A | 390 | S>L | No | 1000Genomes | |
| ENSVATH02956536 | 392 | P>Q | No | 1000Genomes | |
| ENSVATH06822349 | 393 | S>T | No | 1000Genomes | |
| ENSVATH06822348 | 403 | P>L | No | 1000Genomes | |
| ENSVATH06822348 | 403 | P>Q | No | 1000Genomes | |
| ENSVATH02956535 | 405 | A>D | No | 1000Genomes | |
| ENSVATH00551854 | 408 | T>A | No | 1000Genomes | |
| ENSVATH12384667 | 458 | V>I | No | 1000Genomes | |
| tmp_4_16382548_T_C,G | 503 | T>A | No | 1000Genomes | |
| tmp_4_16382548_T_C,G | 503 | T>P | No | 1000Genomes | |
| tmp_4_16382545_C_T | 504 | E>K | No | 1000Genomes | |
| ENSVATH06822347 | 509 | A>V | No | 1000Genomes | |
| tmp_4_16382476_G_A | 527 | P>S | No | 1000Genomes | |
| tmp_4_16382416_C_T | 547 | D>N | No | 1000Genomes | |
| ENSVATH06822345 | 578 | T>N | No | 1000Genomes | |
| ENSVATH06822343 | 598 | E>G | No | 1000Genomes | |
| tmp_4_16382131_T_G | 599 | K>T | No | 1000Genomes | |
| ENSVATH12384633 | 614 | N>T | No | 1000Genomes | |
| tmp_4_16382068_A_G | 620 | I>T | No | 1000Genomes | |
| ENSVATH06822342 | 631 | T>I | No | 1000Genomes | |
| ENSVATH06822341 | 634 | R>P | No | 1000Genomes | |
| ENSVATH06822340 | 636 | S>F | No | 1000Genomes | |
| ENSVATH00551845 | 642 | T>S | No | 1000Genomes | |
| tmp_4_16381983_C_A | 648 | Q>H | No | 1000Genomes | |
| ENSVATH06822339 | 653 | S>A | No | 1000Genomes | |
| ENSVATH14334936 | 664 | V>I | No | 1000Genomes | |
| tmp_4_16381892_C_T | 679 | V>M | No | 1000Genomes | |
| tmp_4_16381855_T_G | 691 | N>T | No | 1000Genomes | |
| tmp_4_16381838_C_T | 697 | A>T | No | 1000Genomes | |
| tmp_4_16381798_G_A | 710 | A>V | No | 1000Genomes | |
| ENSVATH00551844 | 714 | D>N | No | 1000Genomes | |
| ENSVATH02956532 | 730 | I>V | No | 1000Genomes | |
| tmp_4_16381682_C_T | 749 | V>M | No | 1000Genomes | |
| ENSVATH02956531 | 753 | I>F | No | 1000Genomes |
No associated diseases with Q94C77
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to water deprivation | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of deprivation of water. |
| positive regulation of abscisic acid-activated signaling pathway | Any process that activates or increases the frequency, rate or extent of abscisic acid (ABA) signaling. |
| positive regulation of seed germination | Any process that activates or increase the rate of seed germination. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
22 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| O15146 | MUSK | Muscle, skeletal receptor tyrosine-protein kinase | Homo sapiens (Human) | EV |
| P04629 | NTRK1 | High affinity nerve growth factor receptor | Homo sapiens (Human) | EV |
| Q16288 | NTRK3 | NT-3 growth factor receptor | Homo sapiens (Human) | SS |
| Q16620 | NTRK2 | BDNF/NT-3 growth factors receptor | Homo sapiens (Human) | EV |
| P55144 | Tyro3 | Tyrosine-protein kinase receptor TYRO3 | Mus musculus (Mouse) | SS |
| Q61006 | Musk | Muscle, skeletal receptor tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| Q00993 | Axl | Tyrosine-protein kinase receptor UFO | Mus musculus (Mouse) | PR |
| Q60805 | Mertk | Tyrosine-protein kinase Mer | Mus musculus (Mouse) | SS |
| Q3UFB7 | Ntrk1 | High affinity nerve growth factor receptor | Mus musculus (Mouse) | SS |
| P09581 | Csf1r | Macrophage colony-stimulating factor 1 receptor | Mus musculus (Mouse) | SS |
| P21803 | Fgfr2 | Fibroblast growth factor receptor 2 | Mus musculus (Mouse) | SS |
| P16092 | Fgfr1 | Fibroblast growth factor receptor 1 | Mus musculus (Mouse) | SS |
| Q03142 | Fgfr4 | Fibroblast growth factor receptor 4 | Mus musculus (Mouse) | PR |
| P05622 | Pdgfrb | Platelet-derived growth factor receptor beta | Mus musculus (Mouse) | SS |
| Q00342 | Flt3 | Receptor-type tyrosine-protein kinase FLT3 | Mus musculus (Mouse) | SS |
| Q61851 | Fgfr3 | Fibroblast growth factor receptor 3 | Mus musculus (Mouse) | PR |
| P24786 | NTRK3 | NT-3 growth factor receptor | Sus scrofa (Pig) | SS |
| P35739 | Ntrk1 | High affinity nerve growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q03351 | Ntrk3 | NT-3 growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q63604 | Ntrk2 | BDNF/NT-3 growth factors receptor | Rattus norvegicus (Rat) | SS |
| O12990 | jak1 | Tyrosine-protein kinase JAK1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTSNRSNLLF | SLVLFHFLFV | PTQLQALNTD | GVLLLTFKYS | ILTDPLSVLR | NWNYDDATPC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LWTGVTCTEL | GKPNTPDMFR | VTSLVLPNKH | LLGSITPDLF | SIPYLRILDL | SSNFFNGSLP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DSVFNATELQ | SISLGSNNLS | GDLPKSVNSV | TNLQLLNLSA | NAFTGEIPLN | ISLLKNLTVV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SLSKNTFSGD | IPSGFEAAQI | LDLSSNLLNG | SLPKDLGGKS | LHYLNLSHNK | VLGEISPNFA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EKFPANATVD | LSFNNLTGPI | PSSLSLLNQK | AESFSGNQEL | CGKPLKILCS | IPSTLSNPPN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ISETTSPAIA | VKPRSTAPIN | PLTEKPNQTG | KSKLKPSTIA | AITVADIVGL | AFIGLLVLYV |
| 370 | 380 | 390 | 400 | 410 | 420 |
| YQVRKRRRYP | ESSKFSFFKF | CLEKNEAKKS | KPSTTEVTVP | ESPEAKTTCG | SCIILTGGRY |
| 430 | 440 | 450 | 460 | 470 | 480 |
| DETSTSESDV | ENQQTVQAFT | RTDGGQLKQS | SQTQLVTVDG | ETRLDLDTLL | KASAYILGTT |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GTGIVYKAVL | ENGTAFAVRR | IETESCAAAK | PKEFEREVRA | IAKLRHPNLV | RIRGFCWGDD |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EKLLISDYVP | NGSLLCFFTA | TKASSSSSSS | SSLQNPLTFE | ARLKIARGMA | RGLSYINEKK |
| 610 | 620 | 630 | 640 | 650 | 660 |
| QVHGNIKPNN | ILLNAENEPI | ITDLGLDRLM | TPARESHTTG | PTSSSPYQPP | EWSTSLKPNP |
| 670 | 680 | 690 | 700 | 710 | 720 |
| KWDVYSFGVI | LLELLTSKVF | SVDHDIDQFS | NLSDSAAEEN | GRFLRLIDGA | IRSDVARHED |
| 730 | 740 | 750 | |||
| AAMACFRLGI | ECVSSLPQKR | PSMKELVQVL | EKICVLV |