Q92730
Gene name |
RND1 (RHO6) |
Protein name |
Rho-related GTP-binding protein Rho6 |
Names |
Rho family GTPase 1, Rnd1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:27289 |
EC number |
|
Protein Class |
|
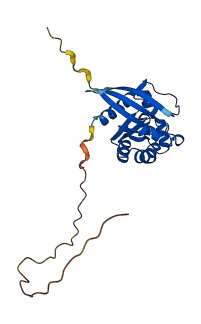
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

4 structures for Q92730
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2CLS | X-ray | 231 A | A/B | 5-200 | PDB |
| 2REX | X-ray | 230 A | B/D | 5-200 | PDB |
| 3Q3J | X-ray | 197 A | B | 5-200 | PDB |
| AF-Q92730-F1 | Predicted | AlphaFoldDB |
160 variants for Q92730
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA6543030 rs140841652 |
3 | E>D | No |
ClinGen ESP ExAC gnomAD |
|
|
CA6543029 rs765113503 |
4 | R>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6543028 rs765113503 |
4 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384640682 rs1592211725 |
4 | R>S | No |
ClinGen Ensembl |
|
|
CA384640674 rs1346869831 |
5 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA6543025 rs760538438 |
6 | A>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6543026 rs753800171 |
6 | A>P | No |
ClinGen ExAC gnomAD |
|
|
CA6543024 rs760538438 |
6 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384640612 rs1290467639 |
7 | P>A | No |
ClinGen gnomAD |
|
|
CA384640575 rs767137867 |
8 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1207127685 CA384640576 |
8 | Q>L | No |
ClinGen TOPMed |
|
|
rs1207127685 CA384640578 TCGA novel |
8 | Q>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen TOPMed NCI-TCGA |
|
rs1207127685 CA384640577 |
8 | Q>R | No |
ClinGen TOPMed |
|
|
CA384640554 rs1185999045 |
9 | P>A | No |
ClinGen gnomAD |
|
|
rs1188397698 CA384640537 |
10 | V>I | No |
ClinGen TOPMed |
|
|
CA384640475 rs1434922959 |
11 | V>A | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs773871104 CA6543020 |
11 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA384640466 rs1471935783 |
12 | A>T | No |
ClinGen TOPMed |
|
|
CA6543019 rs768021613 |
13 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA236603270 rs920395714 |
14 | C>R | No |
ClinGen TOPMed gnomAD |
|
|
CA384640275 rs1254998824 |
20 | G>W | No |
ClinGen gnomAD |
|
|
CA384640260 rs1210398528 |
21 | D>H | No |
ClinGen gnomAD |
|
|
CA384640222 rs1565684340 |
22 | V>L | No |
ClinGen Ensembl |
|
|
rs1325073003 CA384640201 |
23 | Q>E | No |
ClinGen gnomAD |
|
|
CA384640179 rs767999015 |
23 | Q>P | No |
ClinGen TOPMed |
|
|
CA236603268 rs767999015 |
23 | Q>R | No |
ClinGen TOPMed |
|
|
rs746167437 CA6543015 |
28 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA384640059 rs781287162 CA6543014 |
29 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1394019149 CA384640037 |
30 | L>F | No |
ClinGen gnomAD |
|
|
CA6543013 rs757456160 |
31 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA384640025 rs1401251844 |
31 | Q>K | No |
ClinGen gnomAD |
|
|
rs1348068971 CA384639984 |
32 | V>M | No |
ClinGen TOPMed |
|
|
rs1471116750 CA384639899 |
34 | A>V | No |
ClinGen gnomAD |
|
|
rs940626016 CA236603267 |
38 | Y>C | No |
ClinGen Ensembl |
|
|
rs1244778912 CA384638652 |
42 | Y>C | No |
ClinGen TOPMed |
|
|
CA384638597 rs1400896638 |
43 | V>G | No |
ClinGen gnomAD |
|
| TCGA novel | 43 | V>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA6542979 VAR_020188 rs2270577 |
44 | P>R | No |
ClinGen UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
rs1395870529 CA384638447 |
48 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA384638293 rs1201490985 |
51 | T>R | No |
ClinGen gnomAD |
|
|
rs1262169341 CA384638278 |
52 | A>D | No |
ClinGen TOPMed gnomAD |
|
|
rs772370188 CA6542976 |
52 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1262169341 CA384638272 |
52 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1411024373 CA384638262 |
53 | C>Y | No |
ClinGen TOPMed |
|
|
CA384637964 rs1448546419 |
60 | R>S | No |
ClinGen gnomAD |
|
|
rs779173294 CA6542974 |
62 | E>D | No |
ClinGen ExAC |
|
|
rs1324370794 CA384637766 |
65 | L>F | No |
ClinGen gnomAD |
|
|
rs1443866593 CA384637034 |
71 | S>C | No |
ClinGen gnomAD |
|
|
CA6542949 rs747860800 |
75 | D>E | No |
ClinGen ExAC gnomAD |
|
|
COSM1223872 CA384636869 rs1164848675 |
75 | D>N | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
rs1411330184 CA384636832 |
76 | N>D | No |
ClinGen TOPMed gnomAD |
|
|
CA384636745 rs1414211040 |
78 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA236602931 rs551476838 |
78 | R>H | No |
ClinGen TOPMed |
|
|
CA384636710 rs1162476145 |
79 | P>L | No |
ClinGen gnomAD |
|
|
rs527583871 CA6542948 |
80 | L>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA384636585 rs1565683582 |
82 | Y>C | No |
ClinGen Ensembl |
|
|
CA384636568 rs1245144055 |
83 | S>R | No |
ClinGen gnomAD |
|
|
CA6542946 rs753237328 |
84 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1400874748 CA384636481 COSM939922 |
85 | S>L | Variant assessed as Somatic; impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
CA384636448 rs1565683572 |
86 | D>V | No |
ClinGen Ensembl |
|
|
rs756619239 CA6542944 |
88 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs1207223987 CA384636416 |
88 | V>L | No |
ClinGen gnomAD |
|
|
CA384636357 rs1247116893 |
91 | C>R | No |
ClinGen gnomAD |
|
|
rs574042643 CA6542943 |
92 | F>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA6542942 rs145288407 |
94 | I>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6542941 rs145288407 |
94 | I>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs774720622 CA6542940 |
96 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs764356367 CA6542939 |
96 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1298303254 CA384636038 |
98 | E>D | No |
ClinGen gnomAD |
|
|
rs775395215 CA384635919 |
102 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6542936 rs769890484 |
103 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA384635877 rs1424997574 |
104 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1592210505 CA384635848 |
105 | K>Q | No |
ClinGen Ensembl |
|
|
CA384635762 rs1235439433 |
106 | K>R | No |
ClinGen TOPMed |
|
|
rs1453280482 CA384634536 |
107 | W>* | No |
ClinGen gnomAD |
|
|
rs755636649 CA6542924 |
110 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA6542923 rs751037776 |
111 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384634352 rs1424636823 |
115 | C>Y | No |
ClinGen gnomAD |
|
| TCGA novel | 117 | S>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768071716 CA6542922 |
117 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs370916812 CA6542920 |
119 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs370916812 CA6542921 |
119 | R>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6542918 rs144662472 |
119 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs144662472 CA6542919 |
119 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA236602843 rs377652733 |
120 | V>I | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs775762543 CA6542917 |
121 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384634156 rs1217502727 |
122 | L>F | No |
ClinGen gnomAD |
|
|
rs765295653 CA384634129 |
123 | I>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA384634125 rs1282149184 |
123 | I>T | No |
ClinGen gnomAD |
|
|
rs765295653 CA6542916 |
123 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA384633973 rs1431848234 |
125 | C>F | No |
ClinGen TOPMed |
|
|
rs1175778744 CA384633963 |
126 | K>Q | No |
ClinGen TOPMed |
|
|
CA6542915 rs759541705 |
126 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384633831 rs1470711563 |
130 | R>* | No |
ClinGen TOPMed |
|
|
COSM939920 CA6542914 rs267603489 |
130 | R>Q | Variant assessed as Somatic; 0.0 impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
| TCGA novel | 132 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs761624837 CA6542912 |
134 | S>C | No |
ClinGen ExAC gnomAD |
|
|
rs774169802 CA384633438 |
140 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774169802 CA6542911 |
140 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs548432311 CA6542910 |
142 | Q>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs148733686 CA6542908 |
145 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs769192944 CA6542907 |
146 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1386072787 CA384633228 |
146 | P>S | No |
ClinGen TOPMed |
|
|
rs1429415333 CA384633144 |
147 | I>T | No |
ClinGen gnomAD |
|
|
rs200340902 CA6542906 |
149 | Y>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1482097870 CA384632995 |
151 | Q>E | No |
ClinGen gnomAD |
|
|
rs1261928223 CA384632962 |
151 | Q>H | No |
ClinGen gnomAD |
|
|
rs1361527680 CA384631844 |
154 | A>V | No |
ClinGen gnomAD |
|
|
rs1034201641 CA236602571 |
155 | I>T | No |
ClinGen Ensembl |
|
|
CA384631823 rs1440169950 |
156 | A>G | No |
ClinGen gnomAD |
|
|
rs756182208 CA6542877 |
156 | A>P | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 156 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA6542876 rs751456316 |
157 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1463063224 CA384631785 |
158 | Q>H | No |
ClinGen TOPMed |
|
|
rs1319211565 CA384631771 |
160 | G>S | No |
ClinGen TOPMed |
|
|
rs1388465833 CA384631761 |
160 | G>V | No |
ClinGen TOPMed |
|
|
rs764035056 CA6542875 |
161 | A>G | No |
ClinGen ExAC TOPMed |
|
|
rs1252510894 CA384631758 |
161 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA384631749 rs764035056 |
161 | A>V | No |
ClinGen ExAC TOPMed |
|
|
rs1280162358 CA384631744 |
162 | E>Q | No |
ClinGen gnomAD |
|
|
rs1312961279 CA384631724 |
163 | I>V | No |
ClinGen TOPMed |
|
|
CA6542874 rs762679892 |
167 | G>A | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 167 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1224978891 CA384631498 |
175 | S>G | No |
ClinGen TOPMed gnomAD |
|
|
rs760955311 CA6542872 |
178 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs545099199 CA6542871 |
179 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs770565603 CA6542869 |
181 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6542870 COSM1705558 rs144289367 |
181 | R>W | Variant assessed as Somatic; 0.0 impact. skin [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA6542868 rs746520280 |
182 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA236602570 rs930404930 |
182 | T>S | No |
ClinGen TOPMed |
|
|
rs1305387114 CA384631294 |
184 | S>C | No |
ClinGen gnomAD |
|
|
rs1418289451 CA384631267 |
185 | M>T | No |
ClinGen gnomAD |
|
|
rs772604279 CA6542866 |
189 | N>I | No |
ClinGen ExAC gnomAD |
|
|
CA384631009 rs1476228949 |
194 | L>P | No |
ClinGen TOPMed |
|
|
CA6542861 rs780250169 |
195 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs112776514 CA6542862 |
195 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs112776514 CA236602569 |
195 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA384630942 rs1269293757 |
196 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs756243824 CA6542860 |
198 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs750493036 CA6542859 |
198 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs528408574 CA236602568 |
199 | P>S | No |
ClinGen gnomAD |
|
|
CA384630850 rs528408574 |
199 | P>T | No |
ClinGen gnomAD |
|
|
rs149847130 CA6542858 |
200 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1254393474 COSM189969 CA384630792 |
201 | R>* | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA6542857 rs200101480 |
201 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs200101480 CA6542856 |
201 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1356902097 CA384630639 |
206 | R>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA236602566 rs766112027 |
209 | H>Q | No |
ClinGen ExAC TOPMed |
|
|
CA236602567 rs921423708 |
209 | H>Y | No |
ClinGen TOPMed |
|
|
rs1326215617 CA384630477 |
212 | S>G | No |
ClinGen gnomAD |
|
|
rs554533556 CA6542851 |
213 | R>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA6542850 rs772813001 |
213 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs554533556 CA236602565 |
213 | R>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs774774194 CA6542847 |
221 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs564398063 CA236602561 |
222 | K>R | No |
ClinGen Ensembl |
|
|
CA6542846 rs769075692 |
226 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA6542845 rs749638791 |
226 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA236602560 rs977858545 |
229 | C>G | No |
ClinGen Ensembl |
|
|
CA6542842 rs371547164 |
231 | I>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 233 | M>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with Q92730
11 regional properties for Q92730
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Zinc finger C2H2-type | 266 - 293 | IPR013087-1 |
| domain | Zinc finger C2H2-type | 294 - 321 | IPR013087-2 |
| domain | Zinc finger C2H2-type | 322 - 350 | IPR013087-3 |
| domain | Zinc finger C2H2-type | 351 - 378 | IPR013087-4 |
| domain | Zinc finger C2H2-type | 379 - 406 | IPR013087-5 |
| domain | Zinc finger C2H2-type | 407 - 435 | IPR013087-6 |
| domain | Zinc finger C2H2-type | 437 - 465 | IPR013087-7 |
| domain | Zinc finger C2H2-type | 467 - 494 | IPR013087-8 |
| domain | Zinc finger C2H2-type | 495 - 522 | IPR013087-9 |
| domain | Zinc finger C2H2-type | 523 - 546 | IPR013087-10 |
| domain | Zinc finger C2H2-type | 555 - 575 | IPR013087-11 |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| adherens junction | A cell-cell junction composed of the epithelial cadherin-catenin complex. The epithelial cadherins, or E-cadherins, of each interacting cell extend through the plasma membrane into the extracellular space and bind to each other. The E-cadherins bind to catenins on the cytoplasmic side of the membrane, where the E-cadherin-catenin complex binds to cytoskeletal components and regulatory and signaling molecules. |
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| cytoplasmic vesicle | A vesicle found in the cytoplasm of a cell. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cortical cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures in the cell cortex, i.e. just beneath the plasma membrane. |
| establishment or maintenance of cell polarity | Any cellular process that results in the specification, formation or maintenance of anisotropic intracellular organization or cell growth patterns. |
| negative regulation of cell adhesion | Any process that stops, prevents, or reduces the frequency, rate or extent of cell adhesion. |
| neuron remodeling | The developmentally regulated remodeling of neuronal projections such as pruning to eliminate the extra dendrites and axons projections set up in early stages of nervous system development. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| small GTPase mediated signal transduction | The series of molecular signals in which a small monomeric GTPase relays a signal. |
37 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q00246 | RHO4 | GTP-binding protein RHO4 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P61585 | RHOA | Transforming protein RhoA | Bos taurus (Bovine) | PR |
| Q1RMJ6 | RHOC | Rho-related GTP-binding protein RhoC | Bos taurus (Bovine) | PR |
| Q17QI8 | Rhov | Rho-related GTP-binding protein RhoV | Bos taurus (Bovine) | PR |
| Q3ZBW5 | RHOB | Rho-related GTP-binding protein RhoB | Bos taurus (Bovine) | PR |
| Q2HJ68 | RND1 | Rho-related GTP-binding protein Rho6 | Bos taurus (Bovine) | PR |
| Q9PSX7 | RHOC | Rho-related GTP-binding protein RhoC | Gallus gallus (Chicken) | PR |
| P48148 | Rho1 | Ras-like GTP-binding protein Rho1 | Drosophila melanogaster (Fruit fly) | PR |
| O94955 | RHOBTB3 | Rho-related BTB domain-containing protein 3 | Homo sapiens (Human) | EV |
| O94844 | RHOBTB1 | Rho-related BTB domain-containing protein 1 | Homo sapiens (Human) | PR |
| P08134 | RHOC | Rho-related GTP-binding protein RhoC | Homo sapiens (Human) | PR |
| P61586 | RHOA | Transforming protein RhoA | Homo sapiens (Human) | PR |
| P61587 | RND3 | Rho-related GTP-binding protein RhoE | Homo sapiens (Human) | PR |
| P62745 | RHOB | Rho-related GTP-binding protein RhoB | Homo sapiens (Human) | PR |
| Q96L33 | RHOV | Rho-related GTP-binding protein RhoV | Homo sapiens (Human) | PR |
| Q62159 | Rhoc | Rho-related GTP-binding protein RhoC | Mus musculus (Mouse) | PR |
| Q9QUI0 | Rhoa | Transforming protein RhoA | Mus musculus (Mouse) | PR |
| Q9DAK3 | Rhobtb1 | Rho-related BTB domain-containing protein 1 | Mus musculus (Mouse) | PR |
| Q9CTN4 | Rhobtb3 | Rho-related BTB domain-containing protein 3 | Mus musculus (Mouse) | SS |
| Q8VDU1 | Rhov | Rho-related GTP-binding protein RhoV | Mus musculus (Mouse) | PR |
| P62746 | Rhob | Rho-related GTP-binding protein RhoB | Mus musculus (Mouse) | PR |
| P61588 | Rnd3 | Rho-related GTP-binding protein RhoE | Mus musculus (Mouse) | PR |
| Q8BLR7 | Rnd1 | Rho-related GTP-binding protein Rho6 | Mus musculus (Mouse) | PR |
| O77683 | RND3 | Rho-related GTP-binding protein RhoE | Sus scrofa (Pig) | PR |
| P62747 | Rhob | Rho-related GTP-binding protein RhoB | Rattus norvegicus (Rat) | PR |
| Q9Z1Y0 | Rhov | Rho-related GTP-binding protein RhoV | Rattus norvegicus (Rat) | PR |
| P61589 | Rhoa | Transforming protein RhoA | Rattus norvegicus (Rat) | PR |
| Q6SA80 | Rnd3 | Rho-related GTP-binding protein RhoE | Rattus norvegicus (Rat) | PR |
| Q9SSX0 | RAC1 | Rac-like GTP-binding protein 1 | Oryza sativa subsp japonica (Rice) | PR |
| Q6Z808 | RAC3 | Rac-like GTP-binding protein 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q68Y52 | RAC2 | Rac-like GTP-binding protein 2 | Oryza sativa subsp japonica (Rice) | PR |
| Q67VP4 | RAC4 | Rac-like GTP-binding protein 4 | Oryza sativa subsp japonica (Rice) | PR |
| Q22038 | rho-1 | Ras-like GTP-binding protein rhoA | Caenorhabditis elegans | PR |
| Q9SU67 | ARAC8 | Rac-like GTP-binding protein ARAC8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82480 | ARAC7 | Rac-like GTP-binding protein ARAC7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38903 | ARAC2 | Rac-like GTP-binding protein ARAC2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O82481 | ARAC10 | Rac-like GTP-binding protein ARAC10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKERRAPQPV | VARCKLVLVG | DVQCGKTAML | QVLAKDCYPE | TYVPTVFENY | TACLETEEQR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VELSLWDTSG | SPYYDNVRPL | CYSDSDAVLL | CFDISRPETV | DSALKKWRTE | ILDYCPSTRV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LLIGCKTDLR | TDLSTLMELS | HQKQAPISYE | QGCAIAKQLG | AEIYLEGSAF | TSEKSIHSIF |
| 190 | 200 | 210 | 220 | 230 | |
| RTASMLCLNK | PSPLPQKSPV | RSLSKRLLHL | PSRSELISST | FKKEKAKSCS | IM |