Q92519
Gene name |
TRIB2 |
Protein name |
Tribbles homolog 2 |
Names |
TRB-2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:28951 |
EC number |
|
Protein Class |
|
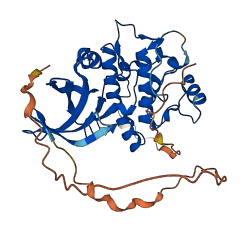
Descriptions
TRIB2 regulates the activity of MAP kinases, and contains a pseudokinase domain and a putative COP1-binding motif. In TRIB1, the pseudokinase masks its COP1-binding motif for autoinhibition. In TRIB2, the pseudokinase and COP1-binding motif may interact each other for autoinhibition.
Autoinhibitory domains (AIDs)
Target domain |
325-331 (COP1-binding motif) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
References
- Murphy JM et al. (2015) "Molecular Mechanism of CCAAT-Enhancer Binding Protein Recruitment by the TRIB1 Pseudokinase", Structure (London, England : 1993), 23, 2111-21
- Li YY et al. (2013) "Novel small-molecule AMPK activator orally exerts beneficial effects on diabetic db/db mice", Toxicology and applied pharmacology, 273, 325-34
- Chen L et al. (2009) "Structural insight into the autoinhibition mechanism of AMP-activated protein kinase", Nature, 459, 1146-9
- Mitchelhill KI et al. (1997) "Posttranslational modifications of the 5'-AMP-activated protein kinase beta1 subunit", The Journal of biological chemistry, 272, 24475-9
- Crute BE et al. (1998) "Functional domains of the alpha1 catalytic subunit of the AMP-activated protein kinase", The Journal of biological chemistry, 273, 35347-54
- Handa N et al. (2011) "Structural basis for compound C inhibition of the human AMP-activated protein kinase α2 subunit kinase domain", Acta crystallographica. Section D, Biological crystallography, 67, 480-7
- Amezcua CA et al. (2002) "Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation", Structure (London, England : 1993), 10, 1349-61
- Rutter J et al. (2001) "PAS kinase: an evolutionarily conserved PAS domain-regulated serine/threonine kinase", Proceedings of the National Academy of Sciences of the United States of America, 98, 8991-6
- Pang T et al. (2007) "Conserved alpha-helix acts as autoinhibitory sequence in AMP-activated protein kinase alpha subunits", The Journal of biological chemistry, 282, 495-506
- Oakhill JS et al. (2009) "Structure and function of AMP-activated protein kinase", Acta physiologica (Oxford, England), 196, 3-14
- Pang T et al. (2008) "Small molecule antagonizes autoinhibition and activates AMP-activated protein kinase in cells", The Journal of biological chemistry, 283, 16051-60
- Scott JW et al. (2014) "ATP sensitive bi-quinoline activator of the AMP-activated protein kinase", Biochemical and biophysical research communications, 443, 435-40
- Beullens M et al. (2005) "Substrate specificity and activity regulation of protein kinase MELK", The Journal of biological chemistry, 280, 40003-11
Autoinhibited structure

Activated structure

2 structures for Q92519
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 7UPM | X-ray | 270 A | A | 53-313 | PDB |
| AF-Q92519-F1 | Predicted | AlphaFoldDB |
199 variants for Q92519
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA345875723 rs1365032723 |
2 | N>S | No |
ClinGen gnomAD |
|
|
CA1534229 rs751185195 |
3 | I>L | No |
ClinGen ExAC gnomAD |
|
|
CA42873670 rs751185195 |
3 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1219495867 CA345875737 |
4 | H>Q | No |
ClinGen gnomAD |
|
|
VAR_042371 rs55813198 CA1534230 |
4 | H>R | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs781024400 CA1534231 |
5 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781024400 CA42873671 |
5 | R>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs371172548 CA1534232 |
6 | S>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs756009177 CA1534233 |
7 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345875749 rs756009177 |
7 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345875758 rs1248688664 |
8 | P>R | No |
ClinGen gnomAD |
|
|
rs1238814895 CA345875777 |
11 | I>T | No |
ClinGen TOPMed |
|
|
rs1281643885 CA345875774 |
11 | I>V | No |
ClinGen TOPMed |
|
|
CA345875780 rs1359455680 |
12 | A>T | No |
ClinGen TOPMed |
|
|
rs1558313620 CA345875819 |
17 | S>L | No |
ClinGen Ensembl |
|
|
rs1395559730 CA345875829 |
19 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA1534236 rs201718135 |
21 | T>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs775750955 CA1534237 |
21 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768775866 CA1534239 |
22 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA345875847 rs1405797074 |
22 | Q>P | No |
ClinGen gnomAD |
|
|
rs1325212906 CA345875852 |
23 | D>N | No |
ClinGen gnomAD |
|
|
rs147226548 CA1534240 |
24 | F>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs765730179 CA42873672 |
26 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1237321106 CA345875885 |
27 | L>S | No |
ClinGen gnomAD |
|
|
CA1534244 rs763495130 |
28 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs200063071 CA1534245 |
28 | S>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345875894 rs1292424309 |
29 | S>P | No |
ClinGen gnomAD |
|
|
rs369189598 CA345875899 |
30 | I>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1534248 rs767077120 |
30 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345875902 rs1213352576 |
30 | I>T | No |
ClinGen gnomAD |
|
|
rs369189598 CA1534247 |
30 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1187498671 CA345875911 |
32 | S>T | No |
ClinGen TOPMed |
|
|
CA345875930 rs755884349 |
34 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1473048566 CA345875933 |
35 | P>S | No |
ClinGen gnomAD |
|
|
COSM1235674 CA1534251 rs531920232 |
36 | S>N | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC gnomAD |
|
rs749206102 CA1534252 |
37 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
CA345875952 rs1572476219 |
38 | S>G | No |
ClinGen Ensembl |
|
|
CA345875974 rs140608739 |
40 | S>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345875978 rs1331701732 |
41 | P>L | No |
ClinGen gnomAD |
|
|
CA345875980 rs1331701732 |
41 | P>R | No |
ClinGen gnomAD |
|
|
CA345875984 rs1572476226 |
42 | N>T | No |
ClinGen Ensembl |
|
|
rs1572476231 CA345875999 |
44 | G>A | No |
ClinGen Ensembl |
|
|
CA345875995 rs1219567868 |
44 | G>R | No |
ClinGen TOPMed |
|
|
CA1534257 rs373415131 |
47 | S>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs768883790 CA1534256 |
47 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA1534259 rs770116295 |
50 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345876046 rs1482195212 |
52 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs763434178 CA1534261 |
53 | N>K | No |
ClinGen ExAC gnomAD |
|
|
CA345876058 rs1259016115 |
54 | L>V | No |
ClinGen gnomAD |
|
|
CA345876068 rs1194158720 |
55 | S>W | No |
ClinGen gnomAD |
|
|
rs1305065428 CA345876075 |
56 | H>L | No |
ClinGen TOPMed |
|
|
CA1534263 rs773779934 |
56 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1014842323 CA42873675 |
58 | V>I | No |
ClinGen TOPMed |
|
|
CA1534264 rs758995072 |
59 | S>T | No |
ClinGen ExAC gnomAD |
|
|
COSM569830 CA1534265 rs767148205 |
62 | G>E | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs755829009 CA345876163 |
69 | P>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM230901 CA1534267 rs755829009 |
69 | P>L | skin [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1171051017 CA345876161 |
69 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1171051017 CA345876160 |
69 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1178642221 CA345876196 |
74 | H>R | No |
ClinGen gnomAD |
|
|
CA345876214 rs1357934399 |
77 | R>C | No |
ClinGen gnomAD |
|
|
rs1027846381 CA42873678 |
77 | R>H | No |
ClinGen TOPMed |
|
|
CA345876224 rs1166829412 |
78 | A>G | No |
ClinGen gnomAD |
|
|
rs1166829412 CA345876223 |
78 | A>V | No |
ClinGen gnomAD |
|
|
rs547834107 CA1534268 |
79 | V>M | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA42873680 rs142544661 |
81 | L>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs142544661 CA1534269 |
81 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345876248 rs375488806 |
82 | H>Q | No |
ClinGen ESP TOPMed |
|
|
CA1534270 rs757196090 |
83 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779026155 CA1534271 |
83 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs754995271 CA1534273 |
86 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA345876286 rs1320313877 |
88 | V>E | No |
ClinGen gnomAD |
|
|
rs781377123 CA1534274 |
90 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA1534297 rs777998116 |
92 | F>S | No |
ClinGen ExAC gnomAD |
|
|
rs749601905 CA1534298 |
94 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1534299 rs576935932 |
100 | S>Y | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs140143213 CA1534300 |
102 | A>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs140143213 CA1534301 |
102 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs377427702 CA1534302 |
106 | C>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs370433974 CA1534303 |
108 | S>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345876440 rs1240954840 |
109 | A>D | No |
ClinGen gnomAD |
|
|
rs1018727454 CA42874185 |
109 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1311451391 CA345876452 |
111 | S>G | No |
ClinGen gnomAD |
|
|
CA345876479 rs1315001044 |
114 | N>S | No |
ClinGen gnomAD |
|
|
CA1534306 rs537841523 |
121 | L>Q | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs765380707 CA1534305 |
121 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA1534307 rs186413267 |
122 | G>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA345876545 rs1488566036 |
124 | T>N | No |
ClinGen gnomAD |
|
|
rs750293522 CA1534309 |
126 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs762743420 CA1534310 |
127 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345876583 rs1370371418 |
130 | F>L | No |
ClinGen TOPMed |
|
|
rs752731552 CA1534312 |
132 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345876642 rs1158862928 |
138 | H>N | No |
ClinGen gnomAD |
|
|
rs1369454287 CA345876655 COSM341253 |
139 | S>F | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA1534315 rs754028360 |
141 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA345876686 rs1253314891 |
144 | C>Y | No |
ClinGen TOPMed |
|
|
rs757541368 CA1534316 |
145 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA345876754 rs1313320132 |
154 | R>K | No |
ClinGen gnomAD |
|
|
CA1534319 rs758793068 |
161 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1200015713 CA345876818 |
164 | A>T | No |
ClinGen gnomAD |
|
|
CA345876829 rs1572478692 |
165 | H>P | No |
ClinGen Ensembl |
|
|
CA345876826 rs1336457667 |
165 | H>Y | No |
ClinGen TOPMed |
|
|
rs769503190 CA1534325 |
168 | D>G | No |
ClinGen ExAC TOPMed |
|
|
CA1534324 rs374295040 |
168 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1534327 rs762835253 |
169 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345876858 rs1163159760 |
169 | G>V | No |
ClinGen gnomAD |
|
|
CA345876875 rs1572478730 |
172 | V>G | No |
ClinGen Ensembl |
|
|
CA1534330 rs759509033 |
174 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA345876889 rs1572478734 |
175 | D>G | No |
ClinGen Ensembl |
|
|
CA345876894 rs1303424574 |
176 | L>F | No |
ClinGen gnomAD |
|
|
CA1534332 rs753974846 |
185 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs546825227 COSM264184 CA1534334 |
186 | E>K | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA1534420 rs372273948 |
190 | R>Q | No |
ClinGen ESP ExAC gnomAD |
|
|
CA1534419 rs749950718 |
190 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1278092834 CA345877437 |
191 | V>I | No |
ClinGen gnomAD |
|
|
CA42876017 rs1004610476 |
194 | E>Q | No |
ClinGen TOPMed |
|
|
CA1534421 rs751324639 |
197 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1191870354 CA345877491 |
199 | A>T | No |
ClinGen gnomAD |
|
|
CA345877507 rs1377749582 |
201 | I>F | No |
ClinGen gnomAD |
|
|
rs748091689 CA1534424 |
203 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781077225 CA1534423 |
203 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1289748767 CA345877520 |
204 | G>R | No |
ClinGen gnomAD |
|
|
CA42876018 rs202225424 |
204 | G>V | No |
ClinGen 1000Genomes |
|
|
rs1380697827 CA345877530 |
205 | D>V | No |
ClinGen gnomAD |
|
|
CA345877552 rs1317864691 |
208 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
CA345877556 rs1295514768 |
209 | L>I | No |
ClinGen TOPMed |
|
|
CA1534427 rs374686543 |
210 | S>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345877567 rs1346973390 COSM1230286 |
211 | D>N | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1239537522 CA345877586 |
213 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1309398763 CA345877610 |
216 | P>L | No |
ClinGen TOPMed |
|
|
CA42876021 rs925303861 |
219 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs762250707 CA1534433 |
226 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345877675 rs762250707 |
226 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1434882386 CA345877683 |
227 | S>N | No |
ClinGen TOPMed |
|
|
CA345877692 rs1365963810 |
228 | G>A | No |
ClinGen gnomAD |
|
|
rs772672172 CA1534435 |
228 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA1534437 rs201044612 |
229 | S>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1323921472 CA345877742 |
236 | D>N | No |
ClinGen gnomAD |
|
|
rs752550457 CA1534441 |
240 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA345877786 rs1572487250 |
242 | V>G | No |
ClinGen Ensembl |
|
|
rs1314980396 CA345877794 |
243 | M>I | No |
ClinGen gnomAD |
|
|
CA1534442 rs756047229 |
246 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs750481302 CA1534444 |
247 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs768938972 CA1534448 |
251 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM1006368 rs186204534 CA1534447 |
251 | R>W | large_intestine endometrium [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
rs1354489788 CA345877878 |
256 | D>G | No |
ClinGen gnomAD |
|
|
CA345877886 rs1181827098 |
257 | I>T | No |
ClinGen gnomAD |
|
|
CA345877918 rs1418882971 |
262 | L>F | No |
ClinGen gnomAD |
|
|
CA345877955 rs1462560406 |
267 | R>Q | No |
ClinGen gnomAD |
|
|
CA1534451 rs770239256 |
267 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA345877959 rs1223287846 |
268 | R>C | No |
ClinGen TOPMed |
|
|
rs980383457 CA42876026 |
268 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs773691736 CA1534452 |
269 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA1534454 rs770337879 |
270 | Q>L | No |
ClinGen ExAC |
|
|
CA345878015 rs773967204 |
276 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs773967204 CA1534455 |
276 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA345878018 rs1558321448 |
277 | L>V | No |
ClinGen Ensembl |
|
|
CA345878027 rs1231128509 |
278 | S>L | No |
ClinGen gnomAD |
|
|
rs1307625146 CA345878034 |
280 | K>Q | No |
ClinGen Ensembl |
|
|
COSM1230288 rs1327493898 CA345878078 |
286 | R>* | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs577789476 COSM1006370 CA1534459 |
286 | R>Q | pancreas endometrium breast [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
CA345878083 rs1253599881 |
287 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
CA1534460 rs764029213 |
290 | R>C | No |
ClinGen ExAC gnomAD |
|
|
COSM1230287 rs1204917469 CA345878103 |
290 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1258712130 CA345878108 |
291 | R>Q | No |
ClinGen gnomAD |
|
|
CA345878116 rs1186167312 |
292 | E>V | No |
ClinGen gnomAD |
|
|
rs202169970 CA42876029 |
296 | R>W | No |
ClinGen 1000Genomes |
|
|
CA345878159 COSM99025 rs1558321486 |
299 | S>L | stomach [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA345878165 rs1429218073 |
300 | Q>R | No |
ClinGen gnomAD |
|
|
CA42876030 rs751664816 |
301 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1352719630 CA345878217 |
307 | W>C | No |
ClinGen gnomAD |
|
|
rs1572487350 CA345878220 |
308 | F>V | No |
ClinGen Ensembl |
|
|
CA345878257 rs1313388768 |
313 | S>N | No |
ClinGen gnomAD |
|
|
COSM1399238 CA1534467 rs376273712 |
314 | V>I | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA1534468 rs770116479 |
315 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1534469 rs770116479 |
315 | S>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA42876031 rs896775225 |
317 | S>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1292755288 CA345878289 |
318 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
CA345878290 rs1226924126 |
319 | Y>H | No |
ClinGen TOPMed |
|
|
CA42876032 rs1014165276 |
320 | G>D | No |
ClinGen TOPMed |
|
|
rs1306386546 CA345878305 |
321 | A>D | No |
ClinGen gnomAD |
|
|
CA42876033 rs1021145140 |
321 | A>P | No |
ClinGen Ensembl |
|
|
CA345878362 rs1572487390 |
329 | V>G | No |
ClinGen Ensembl |
|
|
rs116068311 CA345878376 |
331 | D>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1209106866 CA345878374 |
331 | D>V | No |
ClinGen gnomAD |
|
|
CA1534476 COSM181105 rs760444066 |
332 | V>I | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1413971900 CA345878382 |
333 | N>D | No |
ClinGen TOPMed gnomAD |
|
|
rs763968049 CA1534477 |
333 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA345878396 COSM1494607 rs1558321558 |
334 | M>I | kidney [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA1534478 rs199647863 |
334 | M>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs761590725 COSM181106 CA1534479 |
336 | E>D | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs765253133 CA1534480 |
337 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs1396747166 CA345878420 |
338 | L>V | No |
ClinGen gnomAD |
|
|
CA1534481 rs370540952 |
339 | D>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1803386 CA42876034 |
340 | P>S | No |
ClinGen Ensembl |
|
|
COSM3938497 rs200383586 CA42876035 |
341 | F>L | oesophagus [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA1534483 rs767627435 |
343 | N>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345878455 rs1432515418 |
343 | N>I | No |
ClinGen gnomAD |
|
|
CA1534484 rs752915707 |
344 | N>W | No |
ClinGen ExAC gnomAD |
No associated diseases with Q92519
No regional properties for Q92519
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q92519 | |||
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| mitogen-activated protein kinase kinase binding | Binding to a mitogen-activated protein kinase kinase, a protein that can phosphorylate a MAP kinase. |
| protein kinase inhibitor activity | Binds to and stops, prevents or reduces the activity of a protein kinase, an enzyme which phosphorylates a protein. |
| RNA polymerase II-specific DNA-binding transcription factor binding | Binding to a sequence-specific DNA binding RNA polymerase II transcription factor, any of the factors that interact selectively and non-covalently with a specific DNA sequence in order to modulate transcription. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
| ubiquitin-protein transferase regulator activity | Binds to and modulates the activity of a ubiquitin-protein transferase, an enzyme that catalyzes the covalent attachment of ubiquitin to lysine in a substrate protein. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| negative regulation of fat cell differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of adipocyte differentiation. |
| negative regulation of interleukin-10 production | Any process that stops, prevents, or reduces the frequency, rate, or extent of interleukin-10 production. |
| negative regulation of protein kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of protein kinase activity. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| regulation of MAP kinase activity | Any process that modulates the frequency, rate or extent of MAP kinase activity. |
65 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q02066 | Abscisic acid-inducible protein kinase | Triticum aestivum (Wheat) | PR | |
| Q3SZW1 | TSSK1B | Testis-specific serine/threonine-protein kinase 1 | Bos taurus (Bovine) | PR |
| Q0VCE3 | TRIB3 | Tribbles homolog 3 | Bos taurus (Bovine) | SS |
| Q5GLH2 | TRIB2 | Tribbles homolog 2 | Bos taurus (Bovine) | SS |
| Q28283 | TRIB2 | Tribbles homolog 2 | Canis lupus familiaris (Dog) (Canis familiaris) | SS |
| Q14680 | MELK | Maternal embryonic leucine zipper kinase | Homo sapiens (Human) | EV |
| Q15831 | STK11 | Serine/threonine-protein kinase STK11 | Homo sapiens (Human) | PR |
| Q13131 | PRKAA1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Homo sapiens (Human) | EV |
| Q96RG2 | PASK | PAS domain-containing serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| P54646 | PRKAA2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Homo sapiens (Human) | EV |
| Q9BXA7 | TSSK1B | Testis-specific serine/threonine-protein kinase 1 | Homo sapiens (Human) | PR |
| Q96RU7 | TRIB3 | Tribbles homolog 3 | Homo sapiens (Human) | PR |
| Q96RU8 | TRIB1 | Tribbles homolog 1 | Homo sapiens (Human) | EV |
| Q8BRK8 | Prkaa2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Mus musculus (Mouse) | SS |
| Q61241 | Tssk1b | Testis-specific serine/threonine-protein kinase 1 | Mus musculus (Mouse) | PR |
| Q61846 | Melk | Maternal embryonic leucine zipper kinase | Mus musculus (Mouse) | PR |
| O54863 | Tssk2 | Testis-specific serine/threonine-protein kinase 2 | Mus musculus (Mouse) | PR |
| Q5EG47 | Prkaa1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Mus musculus (Mouse) | SS |
| Q8K4K4 | Trib1 | Tribbles homolog 1 | Mus musculus (Mouse) | SS |
| Q8K4K2 | Trib3 | Tribbles homolog 3 | Mus musculus (Mouse) | SS |
| Q8K4K3 | Trib2 | Tribbles homolog 2 | Mus musculus (Mouse) | SS |
| Q28948 | PRKAA2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Sus scrofa (Pig) | SS |
| Q09137 | Prkaa2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Rattus norvegicus (Rat) | EV |
| P54645 | Prkaa1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Rattus norvegicus (Rat) | EV |
| Q9WTQ6 | Trib3 | Tribbles homolog 3 | Rattus norvegicus (Rat) | SS |
| Q5QNM6 | CIPK13 | Putative CBL-interacting protein kinase 13 | Oryza sativa subsp japonica (Rice) | PR |
| Q5JLQ9 | CIPK30 | CBL-interacting protein kinase 30 | Oryza sativa subsp japonica (Rice) | PR |
| Q5N942 | SAPK4 | Serine/threonine-protein kinase SAPK4 | Oryza sativa subsp japonica (Rice) | PR |
| Q852Q0 | OSK3 | Serine/threonine protein kinase OSK3 | Oryza sativa subsp. japonica (Rice) | SS |
| Q75LR7 | SAPK1 | Serine/threonine-protein kinase SAPK1 | Oryza sativa subsp japonica (Rice) | PR |
| Q75H77 | SAPK10 | Serine/threonine-protein kinase SAPK10 | Oryza sativa subsp japonica (Rice) | PR |
| Q7Y0B9 | SAPK8 | Serine/threonine-protein kinase SAPK8 | Oryza sativa subsp japonica (Rice) | PR |
| Q7XQP4 | SAPK7 | Serine/threonine-protein kinase SAPK7 | Oryza sativa subsp japonica (Rice) | PR |
| Q852Q2 | OSK1 | Serine/threonine protein kinase OSK1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6ZLP5 | CIPK23 | CBL-interacting protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| Q0D4J7 | SAPK2 | Serine/threonine-protein kinase SAPK2 | Oryza sativa subsp japonica (Rice) | PR |
| Q8LIG4 | CIPK3 | CBL-interacting protein kinase 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q852Q1 | OSK4 | Serine/threonine protein kinase OSK4 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6ERS4 | CIPK16 | CBL-interacting protein kinase 16 | Oryza sativa subsp japonica (Rice) | PR |
| P0C5D6 | SAPK3 | Serine/threonine-protein kinase SAPK3 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAX3 | CIPK33 | CBL-interacting protein kinase 33 | Oryza sativa subsp japonica (Rice) | PR |
| Q75V57 | SAPK9 | Serine/threonine-protein kinase SAPK9 | Oryza sativa subsp japonica (Rice) | PR |
| Q21017 | kin-29 | Serine/threonine-protein kinase kin-29 | Caenorhabditis elegans | SS |
| P45894 | aak-1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Caenorhabditis elegans | SS |
| Q95ZQ4 | aak-2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Caenorhabditis elegans | SS |
| G5EED4 | nipi-3 | Protein nipi-3 | Caenorhabditis elegans | PR |
| P43291 | SRK2A | Serine/threonine-protein kinase SRK2A | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q93VD3 | CIPK23 | CBL-interacting serine/threonine-protein kinase 23 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C958 | SRK2B | Serine/threonine-protein kinase SRK2B | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M9E9 | SRK2C | Serine/threonine-protein kinase SRK2C | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64812 | SRK2J | Serine/threonine-protein kinase SRK2J | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q2V452 | CIPK3 | CBL-interacting serine/threonine-protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22971 | CIPK13 | CBL-interacting serine/threonine-protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P92958 | KIN11 | SNF1-related protein kinase catalytic subunit alpha KIN11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39192 | SRK2D | Serine/threonine-protein kinase SRK2D | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O65554 | CIPK6 | CBL-interacting serine/threonine-protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q940H6 | SRK2E | Serine/threonine-protein kinase SRK2E | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P43292 | SRK2G | Serine/threonine-protein kinase SRK2G | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LDI3 | CIPK24 | CBL-interacting serine/threonine-protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FLZ3 | KIN12 | SNF1-related protein kinase catalytic subunit alpha KIN12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FJ54 | CIPK20 | CBL-interacting serine/threonine-protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94CG0 | CIPK21 | CBL-interacting serine/threonine-protein kinase 21 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FFP9 | SRK2H | Serine/threonine-protein kinase SRK2H | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39193 | SRK2I | Serine/threonine-protein kinase SRK2I | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNIHRSTPIT | IARYGRSRNK | TQDFEELSSI | RSAEPSQSFS | PNLGSPSPPE | TPNLSHCVSC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IGKYLLLEPL | EGDHVFRAVH | LHSGEELVCK | VFDISCYQES | LAPCFCLSAH | SNINQITEII |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LGETKAYVFF | ERSYGDMHSF | VRTCKKLREE | EAARLFYQIA | SAVAHCHDGG | LVLRDLKLRK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FIFKDEERTR | VKLESLEDAY | ILRGDDDSLS | DKHGCPAYVS | PEILNTSGSY | SGKAADVWSL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GVMLYTMLVG | RYPFHDIEPS | SLFSKIRRGQ | FNIPETLSPK | AKCLIRSILR | REPSERLTSQ |
| 310 | 320 | 330 | 340 | ||
| EILDHPWFST | DFSVSNSAYG | AKEVSDQLVP | DVNMEENLDP | FFN |