Q91VN6
Gene name |
Ddx41 |
Protein name |
Probable ATP-dependent RNA helicase DDX41 |
Names |
DEAD box protein 41 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:72935 |
EC number |
3.6.4.13: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
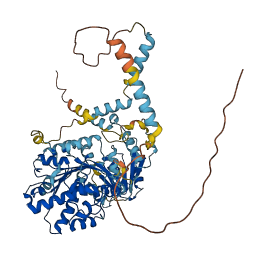
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q91VN6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q91VN6-F1 | Predicted | AlphaFoldDB |
46 variants for Q91VN6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389268840 | 14 | D>V | No | EVA | |
| rs3389284701 | 15 | E>A | No | EVA | |
| rs3389268888 | 15 | E>D | No | EVA | |
| rs3389287891 | 16 | A>S | No | EVA | |
| rs3413000913 | 17 | T>S | No | EVA | |
| rs3389282329 | 19 | G>R | No | EVA | |
| rs3389282593 | 25 | D>G | No | EVA | |
| rs3389296630 | 73 | D>E | No | EVA | |
| rs3389282595 | 86 | S>R | No | EVA | |
| rs3389301875 | 90 | Q>H | No | EVA | |
| rs3389268869 | 101 | R>G | No | EVA | |
| rs3403439370 | 106 | K>C* | No | EVA | |
| rs3389301868 | 125 | A>D | No | EVA | |
| rs3389215084 | 125 | A>S | No | EVA | |
| rs3389215073 | 161 | E>V | No | EVA | |
| rs3389293787 | 164 | R>W | No | EVA | |
| rs3389287944 | 175 | G>D | No | EVA | |
| rs3389282551 | 178 | P>R | No | EVA | |
| rs3389268851 | 180 | I>N | No | EVA | |
| rs3389282572 | 185 | E>D | No | EVA | |
| rs3404304845 | 197 | K>* | No | EVA | |
| rs3389243038 | 206 | P>S | No | EVA | |
| rs3389242923 | 222 | I>V | No | EVA | |
| rs3389301897 | 238 | P>R | No | EVA | |
| rs3389243009 | 254 | K>* | No | EVA | |
| rs3403439407 | 263 | I>F | No | EVA | |
| rs3389287942 | 273 | T>S | No | EVA | |
| rs3389259892 | 296 | L>I | No | EVA | |
| rs3389215063 | 304 | K>N | No | EVA | |
| rs3389287868 | 321 | P>S | No | EVA | |
| rs3389278216 | 386 | A>T | No | EVA | |
| rs3389278183 | 408 | V>I | No | EVA | |
| rs3389284739 | 412 | V>* | No | EVA | |
| rs3389301934 | 434 | P>A | No | EVA | |
| rs3389282244 | 460 | V>I | No | EVA | |
| rs3389215074 | 461 | A>S | No | EVA | |
| rs3389242963 | 461 | A>V | No | EVA | |
| rs3404513571 | 487 | V>E | No | EVA | |
| rs3403501231 | 490 | D>Y | No | EVA | |
| rs3404322606 | 502 | Q>R | No | EVA | |
| rs3389268824 | 511 | E>V | No | EVA | |
| rs3389215051 | 536 | I>N | No | EVA | |
| rs3389259875 | 545 | L>F | No | EVA | |
| rs3411704143 | 577 | G>R | No | EVA | |
| rs3389287691 | 587 | G>S | No | EVA | |
| rs3403977986 | 607 | S>C | No | EVA |
No associated diseases with Q91VN6
25 regional properties for Q91VN6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Toll/interleukin-1 receptor homology (TIR) domain | 865 - 1013 | IPR000157 |
| repeat | Leucine-rich repeat | 123 - 144 | IPR001611-1 |
| repeat | Leucine-rich repeat | 199 - 253 | IPR001611-2 |
| repeat | Leucine-rich repeat | 284 - 343 | IPR001611-3 |
| repeat | Leucine-rich repeat | 475 - 532 | IPR001611-4 |
| repeat | Leucine-rich repeat | 628 - 650 | IPR001611-5 |
| repeat | Leucine-rich repeat | 677 - 698 | IPR001611-6 |
| repeat | Leucine-rich repeat | 701 - 761 | IPR001611-7 |
| repeat | Leucine-rich repeat, typical subtype | 121 - 143 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 197 - 218 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 219 - 241 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 282 - 305 | IPR003591-4 |
| repeat | Leucine-rich repeat, typical subtype | 306 - 331 | IPR003591-5 |
| repeat | Leucine-rich repeat, typical subtype | 362 - 385 | IPR003591-6 |
| repeat | Leucine-rich repeat, typical subtype | 389 - 412 | IPR003591-7 |
| repeat | Leucine-rich repeat, typical subtype | 475 - 493 | IPR003591-8 |
| repeat | Leucine-rich repeat, typical subtype | 494 - 516 | IPR003591-9 |
| repeat | Leucine-rich repeat, typical subtype | 519 - 542 | IPR003591-10 |
| repeat | Leucine-rich repeat, typical subtype | 573 - 597 | IPR003591-11 |
| repeat | Leucine-rich repeat, typical subtype | 626 - 649 | IPR003591-12 |
| repeat | Leucine-rich repeat, typical subtype | 675 - 698 | IPR003591-13 |
| repeat | Leucine-rich repeat, typical subtype | 700 - 722 | IPR003591-14 |
| repeat | Leucine-rich repeat, typical subtype | 724 - 746 | IPR003591-15 |
| repeat | Leucine-rich repeat, typical subtype | 748 - 771 | IPR003591-16 |
| repeat | Leucine-rich repeat unit | 392 - 421 | IPR041283 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.13 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| catalytic step 2 spliceosome | A spliceosomal complex that contains three snRNPs, including U5, bound to a splicing intermediate in which the first catalytic cleavage of the 5' splice site has occurred. The precise subunit composition differs significantly from that of the catalytic step 1, or activated, spliceosome, and includes many proteins in addition to those found in the associated snRNPs. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| spliceosomal complex | Any of a series of ribonucleoprotein complexes that contain snRNA(s) and small nuclear ribonucleoproteins (snRNPs), and are formed sequentially during the spliceosomal splicing of one or more substrate RNAs, and which also contain the RNA substrate(s) from the initial target RNAs of splicing, the splicing intermediate RNA(s), to the final RNA products. During cis-splicing, the initial target RNA is a single, contiguous RNA transcript, whether mRNA, snoRNA, etc., and the released products are a spliced RNA and an excised intron, generally as a lariat structure. During trans-splicing, there are two initial substrate RNAs, the spliced leader RNA and a pre-mRNA. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| metal ion binding | Binding to a metal ion. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| RNA helicase activity | Unwinding of an RNA helix, driven by ATP hydrolysis. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cell population proliferation | The multiplication or reproduction of cells, resulting in the expansion of a cell population. |
| cellular response to interferon-beta | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interferon-beta stimulus. Interferon-beta is a type I interferon. |
| defense response to virus | Reactions triggered in response to the presence of a virus that act to protect the cell or organism. |
| mRNA splicing, via spliceosome | The joining together of exons from one or more primary transcripts of messenger RNA (mRNA) and the excision of intron sequences, via a spliceosomal mechanism, so that mRNA consisting only of the joined exons is produced. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
18 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9V3C0 | abs | ATP-dependent RNA helicase abstrakt | Drosophila melanogaster (Fruit fly) | PR |
| Q9UJV9 | DDX41 | Probable ATP-dependent RNA helicase DDX41 | Homo sapiens (Human) | PR |
| Q91VC3 | Eif4a3 | Eukaryotic initiation factor 4A-III | Mus musculus (Mouse) | PR |
| Q61496 | Ddx4 | ATP-dependent RNA helicase DDX4 | Mus musculus (Mouse) | SS |
| Q62095 | Ddx3y | ATP-dependent RNA helicase DDX3Y | Mus musculus (Mouse) | SS |
| P16381 | D1Pas1 | Putative ATP-dependent RNA helicase Pl10 | Mus musculus (Mouse) | SS |
| Q62167 | Ddx3x | ATP-dependent RNA helicase DDX3X | Mus musculus (Mouse) | SS |
| Q9QY15 | Ddx25 | ATP-dependent RNA helicase DDX25 | Mus musculus (Mouse) | PR |
| Q61655 | Ddx19a | ATP-dependent RNA helicase DDX19A | Mus musculus (Mouse) | SS |
| Q501J6 | Ddx17 | Probable ATP-dependent RNA helicase DDX17 | Mus musculus (Mouse) | PR |
| Q8VDW0 | Ddx39a | ATP-dependent RNA helicase DDX39A | Mus musculus (Mouse) | PR |
| Q9Z1N5 | Ddx39b | Spliceosome RNA helicase Ddx39b | Mus musculus (Mouse) | PR |
| Q99MJ9 | Ddx50 | ATP-dependent RNA helicase DDX50 | Mus musculus (Mouse) | PR |
| Q61656 | Ddx5 | Probable ATP-dependent RNA helicase DDX5 | Mus musculus (Mouse) | PR |
| Q5Z6G5 | Os06g0697200 | DEAD-box ATP-dependent RNA helicase 35B | Oryza sativa subsp japonica (Rice) | PR |
| Q0E3X4 | Os02g0150100 | DEAD-box ATP-dependent RNA helicase 35A | Oryza sativa subsp japonica (Rice) | PR |
| Q9LU46 | RH35 | DEAD-box ATP-dependent RNA helicase 35 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SZB4 | RH43 | Putative DEAD-box ATP-dependent RNA helicase 43 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEDSEPERKR | ARADEATAGG | SRSEDEDEDD | EDYVPYVPLR | QRRQLLLQKL | LQRRRKGATE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EEQQDSGSEP | RGDEDDIPLG | PQSNVSLLDQ | HQHLKEKAEA | RKESAKEKQL | KEEEKILESV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AEGRALMSVK | EMAKGITYDD | PIKTSWTPPR | YVLSMSEERH | ERVRKKYHIL | VEGDGIPPPI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KSFKEMKFPA | AILRGLKKKG | ILHPTPIQIQ | GIPTILSGRD | MIGIAFTGSG | KTLVFTLPVI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| MFCLEQEKRL | PFSKREGPYG | LIICPSRELA | RQTHGILEYY | CRLLQEDSSP | LLRCALCIGG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MSVKEQMETI | RHGVHMMVAT | PGRLMDLLQK | KMVSLDICRY | LALDEADRMI | DMGFEGDIRT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| IFSYFKGQRQ | TLLFSATMPK | KIQNFAKSAL | VKPVTINVGR | AGAASLDVIQ | EVEYVKEEAK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| MVYLLECLQK | TPPPVLIFAE | KKADVDAIHE | YLLLKGVEAV | AIHGGKDQEE | RTKAIEAFRE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GKKDVLVATD | VASKGLDFPA | IQHVINYDMP | EEIENYVHRI | GRTGRSGNTG | IATTFINKAC |
| 550 | 560 | 570 | 580 | 590 | 600 |
| DESVLMDLKA | LLLEAKQKVP | PVLQVLHCGD | ESMLDIGGER | GCAFCGGLGH | RITDCPKLEA |
| 610 | 620 | ||||
| MQTKQVSNIG | RKDYLAHSSM | DF |