Q91987
Gene name |
NTRK2 (TRKB) |
Protein name |
BDNF/NT-3 growth factors receptor |
Names |
EC 2.7.10.1 , Neurotrophic tyrosine kinase receptor type 2 , TrkB tyrosine kinase , Trk-B |
Species |
Gallus gallus (Chicken) |
KEGG Pathway |
gga:396157 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
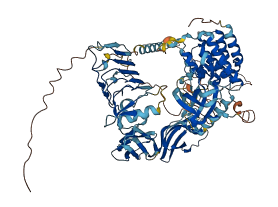
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
534-803 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Target domain |
534-803 (Protein kinase domain) |
Relief mechanism |
Others |
Assay |
|
Accessory elements
689-714 (Activation loop from InterPro)
Target domain |
534-803 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
689-714 (Activation loop from InterPro)
Target domain |
534-803 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Shen J et al. (2019) "Extracellular Juxtamembrane Motif Critical for TrkB Preformed Dimer and Activation", Cells, 8,
- Artim SC et al. (2012) "Assessing the range of kinase autoinhibition mechanisms in the insulin receptor family", The Biochemical journal, 448, 213-20
- Arevalo JC et al. (2000) "TrkA immunoglobulin-like ligand binding domains inhibit spontaneous activation of the receptor", Molecular and cellular biology, 20, 5908-16
- Uchikawa E et al. (2019) "Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex", eLife, 8,
- Nielsen J et al. (2022) "Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling", Journal of molecular biology, 434, 167458
- Chen YS et al. (2021) "Insertion of a synthetic switch into insulin provides metabolite-dependent regulation of hormone-receptor activation", Proceedings of the National Academy of Sciences of the United States of America, 118,
- Craddock BP et al. (2007) "Autoinhibition of the insulin-like growth factor I receptor by the juxtamembrane region", FEBS letters, 581, 3235-40
- Huang X et al. (2009) "Structural insights into the inhibited states of the Mer receptor tyrosine kinase", Journal of structural biology, 165, 88-96
Autoinhibited structure

Activated structure

1 structures for Q91987
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q91987-F1 | Predicted | AlphaFoldDB |
No variants for Q91987
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q91987 | |||||
1 associated diseases with Q91987
[MIM: 613722]: Developmental and epileptic encephalopathy 12 (DEE12)
A form of epilepsy characterized by frequent tonic seizures or spasms beginning in infancy with a specific EEG finding of suppression-burst patterns, characterized by high-voltage bursts alternating with almost flat suppression phases. Patients may progress to West syndrome, which is characterized by tonic spasms with clustering, arrest of psychomotor development, and hypsarrhythmia on EEG. {ECO:0000269|PubMed:20833646}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A form of epilepsy characterized by frequent tonic seizures or spasms beginning in infancy with a specific EEG finding of suppression-burst patterns, characterized by high-voltage bursts alternating with almost flat suppression phases. Patients may progress to West syndrome, which is characterized by tonic spasms with clustering, arrest of psychomotor development, and hypsarrhythmia on EEG. {ECO:0000269|PubMed:20833646}. Note=The disease is caused by variants affecting the gene represented in this entry.
8 regional properties for Q91987
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | C2 domain | 656 - 786 | IPR000008 |
| domain | Phosphatidylinositol-specific phospholipase C, X domain | 316 - 468 | IPR000909 |
| domain | Phospholipase C, phosphatidylinositol-specific, Y domain | 540 - 656 | IPR001711 |
| conserved_site | Phospholipase C-beta, conserved site | 905 - 942 | IPR009535 |
| domain | Phospholipase C-beta, C-terminal domain | 1004 - 1172 | IPR014815 |
| domain | Phosphoinositide-specific phospholipase C, EF-hand-like domain | 216 - 307 | IPR015359 |
| domain | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1, EF hand motif | 153 - 303 | IPR028400 |
| domain | PLC-beta, PH domain | 18 - 148 | IPR037862 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| axon terminus | Terminal inflated portion of the axon, containing the specialized apparatus necessary to release neurotransmitters. The axon terminus is considered to be the whole region of thickening and the terminal button is a specialized region of it. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| endosome membrane | The lipid bilayer surrounding an endosome. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| brain-derived neurotrophic factor binding | Binding to brain-derived neurotrophic factor. |
| brain-derived neurotrophic factor receptor activity | Combining with a brain-derived neurotrophic factor and transmitting the signal across the plasma membrane to initiate a change in cell activity. |
| GPI-linked ephrin receptor activity | Combining with a GPI-anchored ephrin to initiate a change in cell activity. |
| neurotrophin binding | Binding to a neurotrophin, any of a family of growth factors that prevent apoptosis in neurons and promote nerve growth. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| voltage-gated monoatomic ion channel activity | Enables the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons. A voltage-gated channel is a channel whose open state is dependent on the voltage across the membrane in which it is embedded. |
21 GO annotations of biological process
| Name | Definition |
|---|---|
| brain-derived neurotrophic factor receptor signaling pathway | The series of molecular signals generated as a consequence of a brain-derived neurotrophic factor receptor binding to one of its physiological ligands. |
| cellular response to brain-derived neurotrophic factor stimulus | A process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a brain-derived neurotrophic factor stimulus. |
| central nervous system neuron development | The process whose specific outcome is the progression of a neuron whose cell body is located in the central nervous system, from initial commitment of the cell to a neuronal fate, to the fully functional differentiated neuron. |
| cerebral cortex development | The progression of the cerebral cortex over time from its initial formation until its mature state. The cerebral cortex is the outer layered region of the telencephalon. |
| collateral sprouting | The process in which outgrowths develop from the shafts of existing axons. |
| learning | Any process in an organism in which a relatively long-lasting adaptive behavioral change occurs as the result of experience. |
| negative regulation of neuron apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process in neurons. |
| neuron differentiation | The process in which a relatively unspecialized cell acquires specialized features of a neuron. |
| neuron migration | The characteristic movement of an immature neuron from germinal zones to specific positions where they will reside as they mature. |
| positive regulation of axonogenesis | Any process that activates or increases the frequency, rate or extent of axonogenesis. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of collateral sprouting | Any process that activates or increases the frequency, rate or extent of collateral sprouting. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of MAPK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the MAPK cascade. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | Any process that activates or increases the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| regulation of dendrite extension | Any process that modulates the frequency, rate or extent of dendrite extension. |
| regulation of GTPase activity | Any process that modulates the rate of GTP hydrolysis by a GTPase. |
| regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | Any process that modulates the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| trans-synaptic signaling by neuropeptide, modulating synaptic transmission | Cell-cell signaling between presynapse and postsynapse, via the vesicular release and reception of neuropeptide molecules, that modulates the synaptic transmission properties of the synapse. |
41 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q91044 | NTRK3 | NT-3 growth factor receptor | Gallus gallus (Chicken) | SS |
| Q8AXY6 | MUSK | Muscle, skeletal receptor tyrosine protein kinase | Gallus gallus (Chicken) | SS |
| Q5F3X2 | DDR1 | receptor protein-tyrosine kinase | Gallus gallus (Chicken) | SS |
| Q8QHL3 | FLT1 | Vascular endothelial growth factor receptor 1 | Gallus gallus (Chicken) | SS |
| P21804 | FGFR1 | Fibroblast growth factor receptor 1 | Gallus gallus (Chicken) | SS |
| P18461 | FGFR2 | Fibroblast growth factor receptor 2 | Gallus gallus (Chicken) | SS |
| P18460 | FGFR3 | Fibroblast growth factor receptor 3 | Gallus gallus (Chicken) | SS |
| Q98949 | TYRO3 | Tyrosine-protein kinase receptor TYRO3 | Gallus gallus (Chicken) | PR |
| Q08156 | KIT | Mast/stem cell growth factor receptor Kit | Gallus gallus (Chicken) | SS |
| Q91009 | NTRK1 | High affinity nerve growth factor receptor | Gallus gallus (Chicken) | SS |
| Q07494 | EPHB1 | Ephrin type-B receptor 1 | Gallus gallus (Chicken) | SS |
| Q9PUF6 | PDGFRA | Platelet-derived growth factor receptor alpha | Gallus gallus (Chicken) | SS |
| Q5IS37 | NTRK3 | NT-3 growth factor receptor | Pan troglodytes (Chimpanzee) | SS |
| Q24488 | Ror | Tyrosine-protein kinase transmembrane receptor Ror | Drosophila melanogaster (Fruit fly) | SS |
| Q9V6K3 | Nrk | Tyrosine-protein kinase transmembrane receptor Ror2 | Drosophila melanogaster (Fruit fly) | SS |
| Q16288 | NTRK3 | NT-3 growth factor receptor | Homo sapiens (Human) | SS |
| Q01973 | ROR1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Homo sapiens (Human) | PR |
| O15146 | MUSK | Muscle, skeletal receptor tyrosine-protein kinase | Homo sapiens (Human) | EV |
| Q01974 | ROR2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Homo sapiens (Human) | EV |
| P04629 | NTRK1 | High affinity nerve growth factor receptor | Homo sapiens (Human) | EV |
| Q16620 | NTRK2 | BDNF/NT-3 growth factors receptor | Homo sapiens (Human) | EV |
| Q9Z138 | Ror2 | Tyrosine-protein kinase transmembrane receptor ROR2 | Mus musculus (Mouse) | SS |
| Q3UFB7 | Ntrk1 | High affinity nerve growth factor receptor | Mus musculus (Mouse) | SS |
| Q6VNS1 | Ntrk3 | NT-3 growth factor receptor | Mus musculus (Mouse) | SS |
| Q9Z139 | Ror1 | Inactive tyrosine-protein kinase transmembrane receptor ROR1 | Mus musculus (Mouse) | PR |
| Q61006 | Musk | Muscle, skeletal receptor tyrosine-protein kinase | Mus musculus (Mouse) | SS |
| P15209 | Ntrk2 | BDNF/NT-3 growth factors receptor | Mus musculus (Mouse) | SS |
| P24786 | NTRK3 | NT-3 growth factor receptor | Sus scrofa (Pig) | SS |
| P35739 | Ntrk1 | High affinity nerve growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q62838 | Musk | Muscle, skeletal receptor tyrosine protein kinase | Rattus norvegicus (Rat) | SS |
| Q03351 | Ntrk3 | NT-3 growth factor receptor | Rattus norvegicus (Rat) | SS |
| Q63604 | Ntrk2 | BDNF/NT-3 growth factors receptor | Rattus norvegicus (Rat) | SS |
| G5EGK5 | cam-1 | Tyrosine-protein kinase receptor cam-1 | Caenorhabditis elegans | SS |
| C0LGI2 | At1g67720 | Probable LRR receptor-like serine/threonine-protein kinase At1g67720 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGW2 | PAM74 | Probable LRR receptor-like serine/threonine-protein kinase PAM74 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P43298 | TMK1 | Receptor protein kinase TMK1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FN93 | At5g59680 | Probable LRR receptor-like serine/threonine-protein kinase At5g59680 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SNA3 | At3g46340 | Putative receptor-like protein kinase At3g46340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZQR3 | At2g14510 | Leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14510 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64556 | At2g19230 | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g19230 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LFG1 | At3g53590 | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At3g53590 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVSWRRRPGP | GLARLWGLCC | LVLGCWRGAL | GCPASCRCSS | WRIWCSEPVP | GITSFPVPQR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| STEDDNVTEI | YIANQRKLES | INDNEVGFYV | GLKNLTVVDS | GLRFVSRQAF | VKNINLQYIN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LSRNKLSSLS | KKPFRHLGLS | DLILVDNPFK | CSCEIMWIKK | FQETKFYTEA | QDIYCVDDNN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KRIALMDMKV | PNCDLPSANL | SNYNITVVEG | KSITLYCDTT | GGPPPNVSWV | LTNLVSNHES |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DTSKNPASLT | IKNVSSMDSG | LWISCVAENI | VGEVQTSAEL | TVFFAPNITF | IESPTPDHHW |
| 310 | 320 | 330 | 340 | 350 | 360 |
| CIPFTVKGNP | KPTLQWFYEG | AILNESEYIC | TKIHVINQSE | YHGCLQLDNP | THLNNGAYTL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LAKNEYGEDE | KRVDAHFMSV | PGDGSGPIVD | PDVYEYETTP | NDLGDTTNNS | NQITSPDVSN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KENEDSITVY | VVVGIAALVC | TGLVIMLIIL | KFGRHSKFGM | KGPSSVISND | DDSASPLHHI |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SNGSNTPSSS | EGGPDAVIIG | MTKIPVIENP | QYFGITNSQL | KPDTFVQHIK | RHNIVLKREL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GEGAFGKVFL | AECYNLCPEQ | DKILVAVKTL | KDASDNARKD | FHREAELLTN | LQHEHIVKFY |
| 610 | 620 | 630 | 640 | 650 | 660 |
| GVCVEGDPLI | MVFEYMKHGD | LNKFLRAHGP | DAVLMAEGNR | PAELTQSQML | HIAQQIAAGM |
| 670 | 680 | 690 | 700 | 710 | 720 |
| VYLASQHFVH | RDLATRNCLV | GENLLVKIGD | FGMSRDVYST | DYYRVGGHTM | LPIRWMPPES |
| 730 | 740 | 750 | 760 | 770 | 780 |
| IMYRKFTTES | DVWSLGVVLW | EIFTYGKQPW | YQLSNNEVIE | CITQGRVLQR | PRTCPKEVYD |
| 790 | 800 | 810 | |||
| LMLGCWQREP | HMRLNIKEIH | SLLQNLAKAS | PVYLDILG |