Q8VZZ4
Gene name |
ABCC6 (MRP6, MRP8, At3g13090, MJG19.4) |
Protein name |
ABC transporter C family member 6 |
Names |
ABC transporter ABCC.6, AtABCC6, ATP-energized glutathione S-conjugate pump 8, Glutathione S-conjugate-transporting ATPase 8, Multidrug resistance-associated protein 8 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G13090 |
EC number |
7.6.2.2: Linked to the hydrolysis of a nucleoside triphosphate |
Protein Class |
|
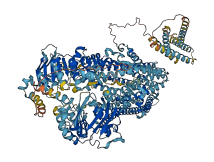
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q8VZZ4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8VZZ4-F1 | Predicted | AlphaFoldDB |
94 variants for Q8VZZ4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_3_4208162_G_A | 4 | P>S | No | 1000Genomes | |
| tmp_3_4208126_A_G | 16 | S>P | No | 1000Genomes | |
| ENSVATH00320358 | 48 | I>V | No | 1000Genomes | |
| ENSVATH10644240 | 57 | S>P | No | 1000Genomes | |
| ENSVATH05819615 | 57 | S>Y | No | 1000Genomes | |
| ENSVATH05819614 | 60 | Y>S | No | 1000Genomes | |
| tmp_3_4207981_A_G | 64 | V>A | No | 1000Genomes | |
| ENSVATH00320357 | 85 | H>Q | No | 1000Genomes | |
| ENSVATH00320356 | 88 | T>I | No | 1000Genomes | |
| tmp_3_4207898_G_C | 92 | P>A | No | 1000Genomes | |
| ENSVATH05819613 | 105 | S>T | No | 1000Genomes | |
| tmp_3_4207801_A_C,T | 124 | F>C | No | 1000Genomes | |
| tmp_3_4207801_A_C,T | 124 | F>Y | No | 1000Genomes | |
| tmp_3_4207791_T_A | 127 | R>S | No | 1000Genomes | |
| tmp_3_4207776_G_C | 132 | F>L | No | 1000Genomes | |
| ENSVATH02138607 | 140 | H>R | No | 1000Genomes | |
| ENSVATH02138606 | 152 | E>K | No | 1000Genomes | |
| ENSVATH05819612 | 156 | V>I | No | 1000Genomes | |
| ENSVATH05819610 | 161 | S>P | No | 1000Genomes | |
| tmp_3_4207679_C_T | 165 | G>S | No | 1000Genomes | |
| tmp_3_4207651_C_A | 174 | C>F | No | 1000Genomes | |
| tmp_3_4207643_A_C | 177 | L>V | No | 1000Genomes | |
| tmp_3_4207620_T_A | 184 | E>D | No | 1000Genomes | |
| ENSVATH00320355 | 200 | S>T | No | 1000Genomes | |
| ENSVATH10644239 | 206 | T>N | No | 1000Genomes | |
| tmp_3_4207553_C_T | 207 | A>T | No | 1000Genomes | |
| ENSVATH10644238 | 238 | K>N | No | 1000Genomes | |
| tmp_3_4207451_G_A | 241 | P>S | No | 1000Genomes | |
| ENSVATH00320354 | 243 | L>V | No | 1000Genomes | |
| ENSVATH05819607 | 272 | F>Y | No | 1000Genomes | |
| ENSVATH05819606 | 309 | N>S | No | 1000Genomes | |
| ENSVATH05819605 | 321 | K>N | No | 1000Genomes | |
| ENSVATH00320353 | 325 | Y>C | No | 1000Genomes | |
| tmp_3_4207151_T_A | 341 | T>S | No | 1000Genomes | |
| tmp_3_4206953_T_C | 407 | I>V | No | 1000Genomes | |
| tmp_3_4206938_C_G | 412 | V>L | No | 1000Genomes | |
| tmp_3_4206935_T_C | 413 | S>G | No | 1000Genomes | |
| ENSVATH13907607 | 427 | S>* | No | 1000Genomes | |
| ENSVATH02138603 | 436 | L>F | No | 1000Genomes | |
| ENSVATH02138602 | 453 | S>G | No | 1000Genomes | |
| ENSVATH05819600 | 458 | S>A | No | 1000Genomes | |
| tmp_3_4206767_C_A | 469 | V>F | No | 1000Genomes | |
| tmp_3_4206708_C_A | 488 | K>N | No | 1000Genomes | |
| ENSVATH05819599 | 502 | K>T | No | 1000Genomes | |
| tmp_3_4206545_C_T | 543 | A>T | No | 1000Genomes | |
| ENSVATH02138599 | 573 | N>I | No | 1000Genomes | |
| ENSVATH02138600 | 573 | N>Y | No | 1000Genomes | |
| tmp_3_4206413_C_T | 587 | D>N | No | 1000Genomes | |
| tmp_3_4206409_A_G | 588 | V>A | No | 1000Genomes | |
| ENSVATH00320352 | 612 | D>E | No | 1000Genomes | |
| tmp_3_4206262_G_C | 637 | T>S | No | 1000Genomes | |
| ENSVATH02138596 | 656 | I>T | No | 1000Genomes | |
| ENSVATH13907564 | 666 | K>N | No | 1000Genomes | |
| tmp_3_4206126_C_A | 682 | E>D | No | 1000Genomes | |
| ENSVATH00320350 | 696 | D>E | No | 1000Genomes | |
| tmp_3_4206048_C_G | 708 | L>F | No | 1000Genomes | |
| tmp_3_4205736_T_A,C | 775 | H>L | No | 1000Genomes | |
| tmp_3_4205736_T_A,C | 775 | H>R | No | 1000Genomes | |
| ENSVATH10644182 | 776 | K>R | No | 1000Genomes | |
| ENSVATH00320347 | 809 | H>N | No | 1000Genomes | |
| ENSVATH02138595 | 830 | A>V | No | 1000Genomes | |
| ENSVATH05819593 | 833 | D>E | No | 1000Genomes | |
| ENSVATH02138594 | 853 | L>I | No | 1000Genomes | |
| ENSVATH02138593 | 856 | K>T | No | 1000Genomes | |
| ENSVATH02138592 | 868 | S>R | No | 1000Genomes | |
| tmp_3_4205364_T_C | 873 | Q>R | No | 1000Genomes | |
| tmp_3_4205248_G_T | 912 | L>I | No | 1000Genomes | |
| ENSVATH10644179 | 939 | T>R | No | 1000Genomes | |
| ENSVATH10644178 | 940 | L>V | No | 1000Genomes | |
| tmp_3_4205106_G_A | 959 | A>V | No | 1000Genomes | |
| ENSVATH13907561 | 997 | I>L | No | 1000Genomes | |
| tmp_3_4204793_C_A | 1027 | L>F | No | 1000Genomes | |
| tmp_3_4204729_C_T | 1049 | A>T | No | 1000Genomes | |
| ENSVATH10644175 | 1056 | Q>H | No | 1000Genomes | |
| ENSVATH02138587 | 1076 | V>M | No | 1000Genomes | |
| ENSVATH05819587 | 1098 | R>M | No | 1000Genomes | |
| tmp_3_4204347_G_A | 1138 | S>F | No | 1000Genomes | |
| tmp_3_4204323_G_A | 1146 | A>V | No | 1000Genomes | |
| ENSVATH05819580 | 1202 | L>F | No | 1000Genomes | |
| tmp_3_4204011_C_A | 1218 | E>D | No | 1000Genomes | |
| ENSVATH07959986 | 1222 | C>Y | No | 1000Genomes | |
| ENSVATH10644103 | 1240 | T>R | No | 1000Genomes | |
| tmp_3_4203916_C_T | 1250 | G>E | No | 1000Genomes | |
| ENSVATH05819578 | 1286 | S>F | No | 1000Genomes | |
| tmp_3_4203704_T_G | 1321 | T>P | No | 1000Genomes | |
| ENSVATH05819577 | 1323 | D>N | No | 1000Genomes | |
| ENSVATH05819576 | 1323 | D>V | No | 1000Genomes | |
| ENSVATH10644101 | 1330 | D>A | No | 1000Genomes | |
| ENSVATH10644100 | 1350 | S>C | No | 1000Genomes | |
| tmp_3_4203427_C_T | 1358 | V>I | No | 1000Genomes | |
| tmp_3_4203302_C_A | 1399 | R>S | No | 1000Genomes | |
| tmp_3_4203217_G_T | 1428 | Q>K | No | 1000Genomes | |
| tmp_3_4203101_G_A | 1438 | A>V | No | 1000Genomes | |
| ENSVATH05819575 | 1460 | E>V | No | 1000Genomes |
No associated diseases with Q8VZZ4
4 regional properties for Q8VZZ4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 36 - 294 | IPR000719 |
| active_site | Serine/threonine-protein kinase, active site | 153 - 165 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 42 - 65 | IPR017441 |
| domain | Serine/threonine-protein kinase 10, catalytic domain | 23 - 314 | IPR042743 |
Functions
| Description | ||
|---|---|---|
| EC Number | 7.6.2.2 | Linked to the hydrolysis of a nucleoside triphosphate |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| plant-type vacuole | A closed structure that is completely surrounded by a unit membrane, contains liquid, and retains the same shape regardless of cell cycle phase. An example of this structure is found in Arabidopsis thaliana. |
| plasmodesma | A fine cytoplasmic channel, found in all higher plants, that connects the cytoplasm of one cell to that of an adjacent cell. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ABC-type xenobiotic transporter activity | Catalysis of the reaction: ATP + H2O + xenobiotic(in) = ADP + phosphate + xenobiotic(out). |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATPase-coupled transmembrane transporter activity | Primary active transporter of a solute across a membrane, via the reaction: ATP + H2O = ADP + phosphate, to directly drive the transport of a substance across a membrane. The transport protein may be transiently phosphorylated (P-type transporters), or not (ABC-type transporters and other families of transporters). Primary active transport occurs up the solute's concentration gradient and is driven by a primary energy source. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| transmembrane transport | The process in which a solute is transported across a lipid bilayer, from one side of a membrane to the other. |
26 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P38735 | VMR1 | ABC transporter ATP-binding protein/permease VMR1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P0CE68 | NFT1 | ABC transporter NFT1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q8HXQ5 | ABCC1 | Multidrug resistance-associated protein 1 | Bos taurus (Bovine) | PR |
| Q2QLE5 | CFTR | Cystic fibrosis transmembrane conductance regulator | Pan troglodytes (Chimpanzee) | PR |
| P91660 | Rh5 | Probable multidrug resistance-associated protein lethal(2)03659 | Drosophila melanogaster (Fruit fly) | PR |
| P13569 | CFTR | Cystic fibrosis transmembrane conductance regulator | Homo sapiens (Human) | PR |
| O95255 | ABCC6 | ATP-binding cassette sub-family C member 6 | Homo sapiens (Human) | PR |
| O15438 | ABCC3 | ATP-binding cassette sub-family C member 3 | Homo sapiens (Human) | PR |
| Q92887 | ABCC2 | ATP-binding cassette sub-family C member 2 | Homo sapiens (Human) | SS |
| O15439 | ABCC4 | ATP-binding cassette sub-family C member 4 | Homo sapiens (Human) | PR |
| P33527 | ABCC1 | Multidrug resistance-associated protein 1 | Homo sapiens (Human) | PR |
| Q96J66 | ABCC11 | ATP-binding cassette sub-family C member 11 | Homo sapiens (Human) | PR |
| B2RX12 | Abcc3 | ATP-binding cassette sub-family C member 3 | Mus musculus (Mouse) | PR |
| Q8VI47 | Abcc2 | ATP-binding cassette sub-family C member 2 | Mus musculus (Mouse) | SS |
| P26361 | Cftr | Cystic fibrosis transmembrane conductance regulator | Mus musculus (Mouse) | PR |
| O35379 | Abcc1 | Multidrug resistance-associated protein 1 | Mus musculus (Mouse) | PR |
| Q80WJ6 | Abcc12 | ATP-binding cassette sub-family C member 12 | Mus musculus (Mouse) | PR |
| Q6PQZ2 | CFTR | Cystic fibrosis transmembrane conductance regulator | Sus scrofa (Pig) | PR |
| Q8CG09 | Abcc1 | Multidrug resistance-associated protein 1 | Rattus norvegicus (Rat) | PR |
| Q63120 | Abcc2 | ATP-binding cassette sub-family C member 2 | Rattus norvegicus (Rat) | EV |
| Q6Y306 | Abcc12 | ATP-binding cassette sub-family C member 12 | Rattus norvegicus (Rat) | PR |
| Q00553 | CFTR | Cystic fibrosis transmembrane conductance regulator | Macaca mulatta (Rhesus macaque) | PR |
| Q9C8G9 | ABCC1 | ABC transporter C family member 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C8H0 | ABCC12 | ABC transporter C family member 12 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M1C7 | ABCC9 | ABC transporter C family member 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SKX0 | ABCC13 | ABC transporter C family member 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MENPIDSLLL | QPIYLSVLSF | FLNLVLLLIL | FGSWLFKKRV | ACEDTDAIMN | EEFKHISFSY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NKLVLICCVS | LSVFYSVLSL | LSCLHWHTNG | WPFLDLLLAA | LTWGSISVYL | FGRYTNSCEQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KVLFLLRVWW | VFFFVVSCYH | LVVDFVLYKK | QEMVSVHFVI | SDLVGVCAGL | FLCCSCLWKK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GEGERIDLLK | EPLLSSAESS | DNEEVTAPFS | KAGILSRMSF | SWMSPLITLG | NEKIIDIKDV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PQLDRSDTTE | SLFWIFRSKL | EWDDGERRIT | TFKLIKALFL | SVWRDIVLSA | LLAFVYTVSC |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YVAPYLMDNF | VQYLNGNRQY | KNQGYVLVTT | FFVAKLVECQ | TQRQWFFRGQ | KAGLGMRSVL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VSMIYEKGLT | LPCHSKQGHT | SGEIINLMAV | DADRISAFSW | FMHDPWILVL | QVSLALWILY |
| 430 | 440 | 450 | 460 | 470 | 480 |
| KSLGLGSIAA | FPATILVMLA | NYPFAKLEEK | FQSSLMKSKD | NRMKKTSEVL | LNMKILKLQG |
| 490 | 500 | 510 | 520 | 530 | 540 |
| WEMKFLSKIL | ELRHIEAGWL | KKFVYNSSAI | NSVLWAAPSF | ISATAFGACL | LLKIPLESGK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ILAALATFRI | LQGPIYKLPE | TISMIVQTKV | SLNRIASFLC | LDDLQQDVVG | RLPSGSSEMA |
| 610 | 620 | 630 | 640 | 650 | 660 |
| VEISNGTFSW | DDSSPIPTLR | DMNFKVSQGM | NVAICGTVGS | GKSSLLSSIL | GEVPKISGNL |
| 670 | 680 | 690 | 700 | 710 | 720 |
| KVCGRKAYIA | QSPWIQSGKV | EENILFGKPM | EREWYDRVLE | ACSLNKDLEI | LPFHDQTVIG |
| 730 | 740 | 750 | 760 | 770 | 780 |
| ERGINLSGGQ | KQRIQIARAL | YQDADIYLFD | DPFSAVDAHT | GSHLFKEVLL | GLLRHKTVIY |
| 790 | 800 | 810 | 820 | 830 | 840 |
| VTHQVEFLPE | ADLILVMKDG | KITQAGKYHE | ILDSGTDFME | LVGAHTEALA | TIDSCETGYA |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SEKSTTDKEN | EVLHHKEKQE | NGSDNKPSGQ | LVQEEEREKG | KVGFTVYKKY | MALAYGGAVI |
| 910 | 920 | 930 | 940 | 950 | 960 |
| PLILVVQVLF | QLLSIGSNYW | MTWVTPVSKD | VEPPVSGFTL | ILVYVLLAVA | SSFCILIRAL |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| LVAMTGFKMA | TELFTQMHLR | IFRASMSFFD | ATPMGRILNR | ASTDQSVADL | RLPGQFAYVA |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| IAAINILGII | GVIVQVAWQV | LIVFIPVVAA | CAWYRQYYIS | AARELARLAG | ISRSPVVHHF |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| SETLSGITTI | RSFDQEPRFR | GDIMRLSDCY | SRLKFHSTGA | MEWLCFRLEL | LSTFAFASSL |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| VILVSAPEGV | INPSLAGLAI | TYALNLNTLQ | ATLIWTLCDL | ENKMISVERM | LQYTNIPSEP |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| PLVIETTRPE | KSWPSRGEIT | ICNLQVRYGP | HLPMVLHGLT | CTFPGGLKTG | IVGRTGCGKS |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| TLIQTLFRIV | EPAAGEIRID | GINILSIGLH | DLRSRLSIIP | QDPTMFEGTI | RSNLDPLEEY |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| TDDQIWEALD | NCQLGDEVRK | KELKLDSPVS | ENGQNWSVGQ | RQLVCLGRVL | LKRSKLLVLD |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| EATASIDTAT | DNLIQETLRH | HFADCTVITI | AHRISSVIDS | DMVLLLDQGL | IKEHDSPARL |
| 1450 | 1460 | ||||
| LEDRSSLFSK | LVAEYTTSSE | SKSKRS |