Q8TAX0
Gene name |
OSR1 (ODD) |
Protein name |
Protein odd-skipped-related 1 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:130497 |
EC number |
|
Protein Class |
|
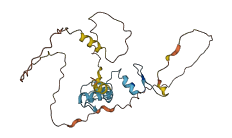
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q8TAX0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8TAX0-F1 | Predicted | AlphaFoldDB |
195 variants for Q8TAX0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1430444346 CA346227690 |
2 | G>D | No |
ClinGen TOPMed |
|
|
CA1542444 rs745601259 |
2 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA346227674 rs1558359994 |
4 | K>N | No |
ClinGen Ensembl |
|
|
CA1542443 rs778633008 |
4 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA346227669 rs1162732154 |
5 | T>S | No |
ClinGen TOPMed |
|
|
CA346227657 rs1415409140 |
7 | P>R | No |
ClinGen gnomAD |
|
| TCGA novel | 7 | P>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs951855533 CA43713585 |
7 | P>T | No |
ClinGen TOPMed |
|
|
rs756917295 CA1542442 |
8 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs1417749076 CA346227639 |
10 | V>A | No |
ClinGen gnomAD |
|
|
CA346227644 rs1158816346 |
10 | V>M | No |
ClinGen gnomAD |
|
|
rs748946826 CA1542441 |
13 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 14 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs777186968 CA1542440 |
14 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1558359952 CA346227611 |
15 | S>A | No |
ClinGen Ensembl |
|
|
CA1542439 rs755578955 |
15 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
CA1542438 rs753222267 |
16 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA346227605 rs753222267 |
16 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767950855 CA346227597 |
17 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755336350 CA1542436 |
20 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA346227569 rs1288940098 |
22 | S>P | No |
ClinGen gnomAD |
|
|
rs530028849 CA1542435 |
25 | Q>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA346227521 rs1297384433 |
29 | G>S | No |
ClinGen gnomAD |
|
|
CA43713517 rs886665270 |
31 | P>R | No |
ClinGen Ensembl |
|
|
CA346227500 rs1436842858 |
33 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA1542433 rs763136719 |
34 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA346227484 rs1323366899 |
35 | S>L | No |
ClinGen gnomAD |
|
|
rs1558359912 CA346227470 |
37 | H>Q | No |
ClinGen Ensembl |
|
|
rs765266778 CA1542431 |
37 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA1542430 rs761936888 |
39 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1542429 rs775583465 |
40 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA346227442 rs1176954593 |
42 | Y>C | No |
ClinGen gnomAD |
|
|
rs1262347464 CA346227436 |
43 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
CA43713500 rs370575190 |
43 | G>S | No |
ClinGen ESP TOPMed gnomAD |
|
| TCGA novel | 46 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs748922957 CA1542424 |
49 | A>P | No |
ClinGen ExAC gnomAD |
|
|
rs748922957 CA346227395 COSM1530150 |
49 | A>S | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA346227396 rs748922957 |
49 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1315057016 CA346227389 |
50 | V>L | No |
ClinGen gnomAD |
|
|
rs1369403119 CA346227377 |
52 | L>M | No |
ClinGen TOPMed gnomAD |
|
|
CA1542422 rs769304282 |
53 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs747587694 CA1542421 |
55 | W>R | No |
ClinGen ExAC gnomAD |
|
|
CA346227344 rs1388802506 |
56 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA346227345 rs1388802506 |
56 | T>R | No |
ClinGen gnomAD |
|
|
CA346227342 rs1455842063 |
57 | L>V | No |
ClinGen gnomAD |
|
|
CA1542419 rs755540358 |
58 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs780326478 CA1542417 |
60 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1542418 rs752087418 |
60 | P>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 61 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1011740676 CA43713458 |
62 | M>R | No |
ClinGen TOPMed |
|
|
CA346227295 rs1321340122 |
64 | L>W | No |
ClinGen gnomAD |
|
|
rs375985128 CA1542415 |
65 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs372422320 CA1542413 |
66 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs372422320 CA1542414 |
66 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA43713449 rs898159433 |
67 | S>A | No |
ClinGen TOPMed |
|
| TCGA novel | 68 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs139101150 CA1542412 |
69 | F>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs868677006 CA346227262 |
70 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
CA43713440 rs868677006 |
70 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
| TCGA novel | 72 | V>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs759585249 CA1542410 |
72 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs947794638 CA43713436 |
73 | P>L | No |
ClinGen gnomAD |
|
|
CA346227245 rs947794638 |
73 | P>Q | No |
ClinGen gnomAD |
|
|
rs774303008 CA1542409 |
73 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA1542407 rs762748716 |
75 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs907421037 CA43713404 |
76 | V>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA1542405 rs769498692 |
78 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1269805691 CA346227209 |
79 | L>F | No |
ClinGen TOPMed |
|
|
CA1542403 rs199763633 |
82 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs199763633 CA43713390 |
82 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1542402 rs148662449 |
83 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM1014239 rs370211451 CA1542401 |
83 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1483660555 CA346227178 |
84 | F>L | No |
ClinGen TOPMed |
|
|
CA43713386 rs140787234 |
87 | P>S | No |
ClinGen ESP TOPMed |
|
|
CA1542399 rs758705032 |
88 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA346227152 rs1572259960 |
89 | F>L | No |
ClinGen Ensembl |
|
|
rs1056276138 CA43713362 |
90 | P>S | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 91 | W>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1542398 rs750733636 |
92 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA1542397 rs779415526 |
93 | P>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA346227121 rs779415526 |
93 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1542396 rs757601212 |
94 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1183484671 COSM1218964 CA346227116 |
94 | H>R | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1486608015 CA346227113 |
95 | V>I | No |
ClinGen gnomAD |
|
|
CA1542395 rs543452352 |
96 | I>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA346227106 rs543452352 |
96 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA346227090 rs1558359786 |
98 | P>R | No |
ClinGen Ensembl |
|
|
CA1542394 rs764299282 |
98 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA346227092 rs764299282 |
98 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1413066323 CA346227078 |
100 | P>S | No |
ClinGen TOPMed |
|
|
CA1542391 rs766269293 |
103 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA346227054 rs1306424843 |
103 | T>I | No |
ClinGen gnomAD |
|
| TCGA novel | 104 | A>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA346227048 rs1299184840 |
104 | A>V | No |
ClinGen gnomAD |
|
|
rs1397194102 CA346227034 |
107 | S>G | No |
ClinGen gnomAD |
|
|
CA346227030 rs1436087381 |
107 | S>N | No |
ClinGen TOPMed |
|
|
rs762792163 CA1542390 |
107 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1369348149 CA346227026 |
108 | V>F | No |
ClinGen TOPMed |
|
|
CA346227012 rs562873645 |
110 | A>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA1542389 rs562873645 COSM2904689 |
110 | A>V | pancreas [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
CA1542388 rs764998863 |
112 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA43713335 rs923800427 |
114 | K>Q | No |
ClinGen gnomAD |
|
|
CA1542386 rs776229107 |
116 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs768179728 CA1542385 |
121 | N>K | No |
ClinGen ExAC |
|
|
rs867715743 CA43713331 |
121 | N>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA43713321 rs769404421 |
124 | L>W | No |
ClinGen Ensembl |
|
| TCGA novel | 126 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA346226906 rs1476174107 |
127 | T>M | No |
ClinGen gnomAD |
|
| TCGA novel | 128 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA346226903 rs1187082054 |
128 | Q>K | No |
ClinGen gnomAD |
|
|
CA1542380 rs779460509 |
128 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1252331658 CA346226895 |
129 | E>Q | No |
ClinGen gnomAD |
|
|
rs1193773342 CA346226877 |
131 | P>R | No |
ClinGen gnomAD |
|
|
rs1191341346 CA346226873 |
132 | A>T | No |
ClinGen gnomAD |
|
|
rs975152724 CA43713311 |
135 | G>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA43713310 rs1018830654 |
135 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs975152724 CA346226853 |
135 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs374867477 CA1542376 |
136 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs374867477 CA1542377 |
136 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA346226847 rs1369401129 |
136 | R>L | No |
ClinGen TOPMed |
|
|
CA346226846 rs767477557 CA1542374 |
137 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 138 | E>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 139 | G>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1542371 rs765050076 |
139 | G>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 140 | P>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1463104651 CA346226819 |
141 | G>D | No |
ClinGen gnomAD |
|
|
CA346226821 rs1294289762 |
141 | G>R | No |
ClinGen gnomAD |
|
|
CA346226822 rs1294289762 |
141 | G>S | No |
ClinGen gnomAD |
|
|
CA1542367 rs760240477 |
142 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1542368 rs760240477 |
142 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1164141110 CA346226802 |
144 | A>E | No |
ClinGen gnomAD |
|
|
CA1542366 rs775124127 |
145 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs1179360144 CA346226793 |
146 | G>E | No |
ClinGen gnomAD |
|
|
rs771451659 CA1542365 |
146 | G>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 146 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1267644701 CA346226783 |
148 | G>C | No |
ClinGen gnomAD |
|
|
CA43713256 rs893197225 |
148 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1267644701 CA346226785 |
148 | G>S | No |
ClinGen gnomAD |
|
|
CA1542363 rs149742000 |
149 | A>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA346226776 rs149742000 |
149 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1263865115 CA346226767 |
151 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1271925561 CA346226761 |
152 | D>G | No |
ClinGen gnomAD |
|
|
CA1542362 rs771434319 |
152 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA346226752 rs1572259744 |
153 | V>G | No |
ClinGen Ensembl |
|
|
CA1542360 rs376388576 |
153 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs142934357 CA346226745 |
155 | K>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs142934357 CA1542359 |
155 | K>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA43713237 rs147259951 |
157 | S>P | No |
ClinGen ESP |
|
| TCGA novel | 159 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1542358 rs574615795 |
159 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA346226714 rs1221411052 |
160 | K>E | No |
ClinGen TOPMed |
|
|
CA346226708 rs1247687590 |
160 | K>N | No |
ClinGen TOPMed |
|
|
CA43713226 rs781377076 |
161 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA43713221 rs963629347 |
162 | P>L | No |
ClinGen TOPMed |
|
|
CA1542355 rs754976969 |
162 | P>S | No |
ClinGen ExAC gnomAD |
|
|
COSM1014238 rs1457744690 CA346226674 |
166 | R>H | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA346226664 rs1156697377 |
168 | P>S | No |
ClinGen gnomAD |
|
| TCGA novel | 170 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 173 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 180 | C>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1404730075 CA346226565 |
181 | G>D | No |
ClinGen TOPMed |
|
|
CA346226554 rs1165317852 |
183 | H>R | No |
ClinGen TOPMed |
|
| TCGA novel | 184 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs752250481 CA1542348 |
190 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1542347 rs767076399 |
192 | I>N | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 193 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1542344 rs771630452 |
199 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA346226448 rs1236849681 |
199 | D>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1236849681 CA346226447 |
199 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1328234056 CA346226410 |
204 | T>I | No |
ClinGen gnomAD |
|
|
rs1402723981 CA346226390 |
207 | I>T | No |
ClinGen gnomAD |
|
|
CA1542340 rs748437826 |
207 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA346226374 rs1337476179 |
209 | H>L | No |
ClinGen TOPMed |
|
|
rs866968142 CA43713160 |
213 | R>Q | No |
ClinGen Ensembl |
|
| TCGA novel | 216 | D>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA346226196 rs1193066952 |
232 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1231313639 CA346226172 |
235 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1572259235 CA346226163 |
236 | C>W | No |
ClinGen Ensembl |
|
| TCGA novel | 241 | C>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 243 | S>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs138448676 CA1542313 |
243 | S>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1257733324 CA346226117 |
243 | S>P | No |
ClinGen TOPMed gnomAD |
|
|
CA346226105 rs1572259211 |
245 | T>P | No |
ClinGen Ensembl |
|
|
CA346226098 rs1279917154 |
246 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA346226063 rs1196801661 |
251 | T>M | No |
ClinGen TOPMed gnomAD |
|
|
CA1542309 rs766022107 |
254 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA346226034 rs1558359236 |
256 | V>M | No |
ClinGen Ensembl |
|
|
rs765599206 CA346226016 |
258 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA1542307 rs149037267 |
258 | E>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs34404071 CA346226020 |
258 | E>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1542308 rs34404071 |
258 | E>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 260 | K>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA43712502 rs555776548 |
264 | I>F | No |
ClinGen gnomAD |
|
|
rs1558359218 CA346225964 |
266 | C>R | No |
ClinGen Ensembl |
|
|
CA346225957 rs1365155354 |
267 | C>Q | No |
ClinGen Ensembl |
No associated diseases with Q8TAX0
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extrinsic component of membrane | The component of a membrane consisting of gene products and protein complexes that are loosely bound to one of its surfaces, but not integrated into the hydrophobic region. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| metal ion binding | Binding to a metal ion. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
51 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cell proliferation involved in kidney development | The multiplication or reproduction of cells, resulting in the expansion of the population in the kidney. |
| cellular response to retinoic acid | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a retinoic acid stimulus. |
| chondrocyte differentiation | The process in which a chondroblast acquires specialized structural and/or functional features of a chondrocyte. A chondrocyte is a polymorphic cell that forms cartilage. |
| embryonic digit morphogenesis | The process, occurring in the embryo, by which the anatomical structures of the digit are generated and organized. A digit is one of the terminal divisions of an appendage, such as a finger or toe. |
| embryonic forelimb morphogenesis | The process, occurring in the embryo, by which the anatomical structures of the forelimb are generated and organized. The forelimbs are the front limbs of an animal, e.g. the arms of a human. |
| embryonic hindlimb morphogenesis | The process, occurring in the embryo, by which the anatomical structures of the hindlimbs are generated and organized. The hindlimbs are the posterior limbs of an animal. |
| embryonic skeletal joint development | The process, occurring during the embryonic phase, whose specific outcome is the progression of the skeletal joints over time, from formation to mature structure. |
| embryonic skeletal joint morphogenesis | The process in which the anatomical structures of skeletal joints are generated and organized during the embryonic phase. A skeletal joint is the connecting structure between the bones of the skeleton. |
| embryonic skeletal limb joint morphogenesis | The process, occurring in the embryo, in which the anatomical structures of a skeletal limb joint are generated and organized. A skeletal limb joint is the connecting structure between the bones of a limb. |
| gonad development | The process whose specific outcome is the progression of the gonad over time, from its formation to the mature structure. The gonad is an animal organ that produces gametes; in some species it also produces hormones. |
| heart development | The process whose specific outcome is the progression of the heart over time, from its formation to the mature structure. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood. |
| intermediate mesoderm development | The process whose specific outcome is the progression of the intermediate mesoderm over time, from its formation to the mature structure. The intermediate mesoderm is located between the lateral mesoderm and the paraxial mesoderm. It develops into the kidney and gonads. |
| mesangial cell development | The process whose specific outcome is the progression of a mesangial cell in the kidney over time, from its formation to the mature structure. |
| mesonephric duct morphogenesis | The process in which the anatomical structures of the mesonephric duct are generated and organized. A mesonephric duct is a tube drains the mesonephros. |
| mesonephros development | The process whose specific outcome is the progression of the mesonephros over time, from its formation to the mature structure. In mammals, the mesonephros is the second of the three embryonic kidneys to be established and exists only transiently. In lower vertebrates such as fish and amphibia, the mesonephros will form the mature kidney. |
| metanephric cap mesenchymal cell proliferation involved in metanephros development | The multiplication or reproduction of metanephric cap mesenchymal cells, resulting in the expansion of the cell population. A metanephric cap mesenchymal cell is a mesenchymal cell that has condensed with other mesenchymal cells surrounding the ureteric bud tip. |
| metanephric epithelium development | The process whose specific outcome is the progression of an epithelium in the metanephros over time, from its formation to the mature structure. An epithelium is a tissue that covers the internal or external surfaces of an anatomical structure. |
| metanephric glomerulus vasculature development | The biological process whose specific outcome is the progression of a metanephric glomerulus vasculature from an initial condition to its mature state. This process begins with the formation of the metanephric glomerulus vasculature and ends with the mature structure. The metanephric glomerulus vasculature is composed of the tubule structures that carry blood or lymph in the metanephric glomerulus. |
| metanephric interstitial fibroblast development | The process whose specific outcome is the progression of a metanephric interstitial fibroblast over time, from its formation to the mature structure. |
| metanephric mesenchymal cell differentiation | The process in which relatively unspecialized cells acquire specialized structural and/or functional features that characterize the mesenchymal cells of the metanephros as it progresses from its formation to the mature state. |
| metanephric mesenchyme development | The biological process whose specific outcome is the progression of a metanephric mesenchyme from an initial condition to its mature state. This process begins with the formation of metanephric mesenchyme and ends with the mature structure. Metanephric mesenchyme is the tissue made up of loosely connected mesenchymal cells in the metanephros. |
| metanephric mesenchyme morphogenesis | The process in which the anatomical structures of a metanephric mesenchymal tissue are generated and organized. Metanephric mesenchyme is the tissue made up of loosely connected mesenchymal cells in the metanephros. |
| metanephric nephron tubule development | The progression of a metanephric nephron tubule over time, from its initial formation to the mature structure. A metanephric nephron tubule is an epithelial tube that is part of the metanephric nephron, the functional part of the metanephros. |
| metanephric smooth muscle tissue development | The process whose specific outcome is the progression of smooth muscle in the metanephros over time, from its formation to the mature structure. |
| middle ear morphogenesis | The process in which the anatomical structures of the middle ear are generated and organized. The middle ear is the air-filled cavity within the skull of vertebrates that lies between the outer ear and the inner ear. It is linked to the pharynx (and therefore to outside air) via the Eustachian tube and in mammals contains the three ear ossicles, which transmit auditory vibrations from the outer ear (via the tympanum) to the inner ear (via the oval window). |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of creatine transmembrane transporter activity | Any process that stops, prevents or reduces the frequency, rate or extent of creatine transmembrane transporter activity. |
| negative regulation of epithelial cell differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of epithelial cell differentiation. |
| negative regulation of nephron tubule epithelial cell differentiation | Any process that decreases the frequency, rate or extent of nephron tubule epithelial cell differentiation. |
| negative regulation of sodium ion transmembrane transporter activity | Any process that stops, prevents or reduces the frequency, rate or extent of sodium ion transmembrane transporter activity. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| odontogenesis | The process whose specific outcome is the progression of a tooth or teeth over time, from formation to the mature structure(s). A tooth is any hard bony, calcareous, or chitinous organ found in the mouth or pharynx of an animal and used in procuring or masticating food. |
| pattern specification involved in metanephros development | Any developmental process that results in the creation of defined areas or spaces within the metanephros to which cells respond and eventually are instructed to differentiate. |
| positive regulation of bone mineralization | Any process that activates or increases the frequency, rate or extent of bone mineralization. |
| positive regulation of epithelial cell proliferation | Any process that activates or increases the rate or extent of epithelial cell proliferation. |
| positive regulation of gastrulation | Any process that activates or increases the frequency, rate or extent of gastrulation. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| posterior mesonephric tubule development | The progression of the posterior mesonephric tubule over time, from its initial formation to the mature structure. The posterior mesonephric tubule is an epithelial tube that is part of the mesonephros. |
| pronephros development | The process whose specific outcome is the progression of the pronephros over time, from its formation to the mature structure. In mammals, the pronephros is the first of the three embryonic kidneys to be established and exists only transiently. In lower vertebrates such as fish and amphibia, the pronephros is the fully functional embryonic kidney and is indispensable for larval life. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| renal vesicle progenitor cell differentiation | The process in which relatively unspecialized cells acquire specialized structural and/or functional features that characterize the renal vesicle progenitor cells of the kidney as it progresses from its formation to the mature state. A renal vesicle progenitor cell is a cell that will give rise to terminally differentiated cells of the renal vesicle without self-renewing. |
| roof of mouth development | The biological process whose specific outcome is the progression of the roof of the mouth from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure. The roof of the mouth is the partition that separates the nasal and oral cavities. |
| sodium ion transmembrane transport | A process in which a sodium ion is transported from one side of a membrane to the other by means of some agent such as a transporter or pore. |
| specification of anterior mesonephric tubule identity | The process in which the tubules of the anterior mesonephros acquire their identity. |
| specification of posterior mesonephric tubule identity | The process in which the tubules of the posterior mesonephros acquire their identity. |
| stem cell differentiation | The process in which a relatively unspecialized cell acquires specialized features of a stem cell. A stem cell is a cell that retains the ability to divide and proliferate throughout life to provide progenitor cells that can differentiate into specialized cells. |
| ureter urothelium development | The process whose specific outcome is the progression of the urothelium of the ureter over time, from its formation to the mature structure. The urothelium is an epithelium that makes up the epithelial tube of the ureter. |
| ureteric bud development | The process whose specific outcome is the progression of the ureteric bud over time, from its formation to the mature structure. |
| urogenital system development | The process whose specific outcome is the progression of the urogenital system over time, from its formation to the mature structure. |
177 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2VWH6 | FEZF2 | Fez family zinc finger protein 2 | Bos taurus (Bovine) | PR |
| A6QNZ0 | ZSCAN26 | Zinc finger and SCAN domain-containing protein 26 | Bos taurus (Bovine) | PR |
| A7MBI1 | ZFP69 | Zinc finger protein 69 homolog | Bos taurus (Bovine) | PR |
| Q08DS3 | OSR1 | Protein odd-skipped-related 1 | Bos taurus (Bovine) | PR |
| Q08705 | CTCF | Transcriptional repressor CTCF | Gallus gallus (Chicken) | PR |
| O42409 | GFI1B | Zinc finger protein Gfi-1b | Gallus gallus (Chicken) | PR |
| A2T6W2 | ZNF449 | Zinc finger protein 449 | Pan troglodytes (Chimpanzee) | PR |
| Q9U405 | grau | Transcription factor grauzone | Drosophila melanogaster (Fruit fly) | PR |
| Q7K0S9 | sug | Zinc finger protein GLIS2 homolog | Drosophila melanogaster (Fruit fly) | PR |
| P20385 | Cf2 | Chorion transcription factor Cf2 | Drosophila melanogaster (Fruit fly) | PR |
| Q86P48 | ATbp | AT-rich binding protein | Drosophila melanogaster (Fruit fly) | PR |
| P28698 | MZF1 | Myeloid zinc finger 1 | Homo sapiens (Human) | PR |
| Q9NTW7 | ZFP64 | Zinc finger protein 64 | Homo sapiens (Human) | PR |
| O14978 | ZNF263 | Zinc finger protein 263 | Homo sapiens (Human) | PR |
| O60304 | ZNF500 | Zinc finger protein 500 | Homo sapiens (Human) | PR |
| P08151 | GLI1 | Zinc finger protein GLI1 | Homo sapiens (Human) | PR |
| Q9UFB7 | ZBTB47 | Zinc finger and BTB domain-containing protein 47 | Homo sapiens (Human) | PR |
| P18146 | EGR1 | Early growth response protein 1 | Homo sapiens (Human) | PR |
| Q9Y5W3 | KLF2 | Krueppel-like factor 2 | Homo sapiens (Human) | PR |
| Q9UNY5 | ZNF232 | Zinc finger protein 232 | Homo sapiens (Human) | PR |
| Q96SZ4 | ZSCAN10 | Zinc finger and SCAN domain-containing protein 10 | Homo sapiens (Human) | PR |
| P17028 | ZNF24 | Zinc finger protein 24 | Homo sapiens (Human) | PR |
| P57682 | KLF3 | Krueppel-like factor 3 | Homo sapiens (Human) | PR |
| P25490 | YY1 | Transcriptional repressor protein YY1 | Homo sapiens (Human) | SS |
| O43296 | ZNF264 | Zinc finger protein 264 | Homo sapiens (Human) | PR |
| P49711 | CTCF | Transcriptional repressor CTCF | Homo sapiens (Human) | PR |
| Q9NQX1 | PRDM5 | PR domain zinc finger protein 5 | Homo sapiens (Human) | PR |
| Q9HBE1 | PATZ1 | POZ-, AT hook-, and zinc finger-containing protein 1 | Homo sapiens (Human) | PR |
| Q9UL58 | ZNF215 | Zinc finger protein 215 | Homo sapiens (Human) | PR |
| Q8TBJ5 | FEZF2 | Fez family zinc finger protein 2 | Homo sapiens (Human) | PR |
| Q96SR6 | ZNF382 | Zinc finger protein 382 | Homo sapiens (Human) | PR |
| Q96IT1 | ZNF496 | Zinc finger protein 496 | Homo sapiens (Human) | PR |
| Q96N95 | ZNF396 | Zinc finger protein 396 | Homo sapiens (Human) | PR |
| Q9ULJ3 | ZBTB21 | Zinc finger and BTB domain-containing protein 21 | Homo sapiens (Human) | PR |
| O75840 | KLF7 | Krueppel-like factor 7 | Homo sapiens (Human) | PR |
| Q9H9D4 | ZNF408 | Zinc finger protein 408 | Homo sapiens (Human) | PR |
| Q13127 | REST | RE1-silencing transcription factor | Homo sapiens (Human) | PR |
| Q8IZM8 | ZNF654 | Zinc finger protein 654 | Homo sapiens (Human) | PR |
| Q14526 | HIC1 | Hypermethylated in cancer 1 protein | Homo sapiens (Human) | PR |
| P17022 | ZNF18 | Zinc finger protein 18 | Homo sapiens (Human) | PR |
| Q86XF7 | ZNF575 | Zinc finger protein 575 | Homo sapiens (Human) | PR |
| Q06889 | EGR3 | Early growth response protein 3 | Homo sapiens (Human) | PR |
| Q8NAM6 | ZSCAN4 | Zinc finger and SCAN domain-containing protein 4 | Homo sapiens (Human) | PR |
| Q08ER8 | ZNF543 | Zinc finger protein 543 | Homo sapiens (Human) | PR |
| P17029 | ZKSCAN1 | Zinc finger protein with KRAB and SCAN domains 1 | Homo sapiens (Human) | PR |
| Q8N680 | ZBTB2 | Zinc finger and BTB domain-containing protein 2 | Homo sapiens (Human) | PR |
| O95625 | ZBTB11 | Zinc finger and BTB domain-containing protein 11 | Homo sapiens (Human) | PR |
| Q9NPC7 | MYNN | Myoneurin | Homo sapiens (Human) | PR |
| Q96BV0 | ZNF775 | Zinc finger protein 775 | Homo sapiens (Human) | PR |
| Q8NF99 | ZNF397 | Zinc finger protein 397 | Homo sapiens (Human) | PR |
| Q63HK3 | ZKSCAN2 | Zinc finger protein with KRAB and SCAN domains 2 | Homo sapiens (Human) | PR |
| Q5FWF6 | ZNF789 | Zinc finger protein 789 | Homo sapiens (Human) | PR |
| Q15776 | ZKSCAN8 | Zinc finger protein with KRAB and SCAN domains 8 | Homo sapiens (Human) | PR |
| Q53GI3 | ZNF394 | Zinc finger protein 394 | Homo sapiens (Human) | PR |
| O95125 | ZNF202 | Zinc finger protein 202 | Homo sapiens (Human) | PR |
| Q05516 | ZBTB16 | Zinc finger and BTB domain-containing protein 16 | Homo sapiens (Human) | PR |
| Q9H116 | GZF1 | GDNF-inducible zinc finger protein 1 | Homo sapiens (Human) | PR |
| Q8N0Y2 | ZNF444 | Zinc finger protein 444 | Homo sapiens (Human) | PR |
| Q6P9G9 | ZNF449 | Zinc finger protein 449 | Homo sapiens (Human) | PR |
| Q8IW36 | ZNF695 | Zinc finger protein 695 | Homo sapiens (Human) | PR |
| Q5VTD9 | GFI1B | Zinc finger protein Gfi-1b | Homo sapiens (Human) | PR |
| Q6PG37 | ZNF790 | Zinc finger protein 790 | Homo sapiens (Human) | PR |
| Q9NQV6 | PRDM10 | PR domain zinc finger protein 10 | Homo sapiens (Human) | PR |
| Q9Y2D9 | ZNF652 | Zinc finger protein 652 | Homo sapiens (Human) | PR |
| Q5TC79 | ZBTB37 | Zinc finger and BTB domain-containing protein 37 | Homo sapiens (Human) | PR |
| Q9Y4E5 | ZNF451 | E3 SUMO-protein ligase ZNF451 | Homo sapiens (Human) | PR |
| Q8ND82 | ZNF280C | Zinc finger protein 280C | Homo sapiens (Human) | PR |
| Q49AA0 | ZFP69 | Zinc finger protein 69 homolog | Homo sapiens (Human) | PR |
| O43298 | ZBTB43 | Zinc finger and BTB domain-containing protein 43 | Homo sapiens (Human) | PR |
| Q9Y330 | ZBTB12 | Zinc finger and BTB domain-containing protein 12 | Homo sapiens (Human) | PR |
| Q13105 | ZBTB17 | Zinc finger and BTB domain-containing protein 17 | Homo sapiens (Human) | PR |
| P51508 | ZNF81 | Zinc finger protein 81 | Homo sapiens (Human) | PR |
| Q5JNZ3 | ZNF311 | Zinc finger protein 311 | Homo sapiens (Human) | PR |
| Q9BRR0 | ZKSCAN3 | Zinc finger protein with KRAB and SCAN domains 3 | Homo sapiens (Human) | PR |
| Q969J2 | ZKSCAN4 | Zinc finger protein with KRAB and SCAN domains 4 | Homo sapiens (Human) | PR |
| P49910 | ZNF165 | Zinc finger protein 165 | Homo sapiens (Human) | PR |
| Q9Y4X4 | KLF12 | Krueppel-like factor 12 | Homo sapiens (Human) | PR |
| P10074 | ZBTB48 | Telomere zinc finger-associated protein | Homo sapiens (Human) | PR |
| P17010 | ZFX | Zinc finger X-chromosomal protein | Homo sapiens (Human) | PR |
| Q9H5H4 | ZNF768 | Zinc finger protein 768 | Homo sapiens (Human) | PR |
| Q6NSZ9 | ZSCAN25 | Zinc finger and SCAN domain-containing protein 25 | Homo sapiens (Human) | PR |
| Q9Y2L8 | ZKSCAN5 | Zinc finger protein with KRAB and SCAN domains 5 | Homo sapiens (Human) | PR |
| Q86UZ6 | ZBTB46 | Zinc finger and BTB domain-containing protein 46 | Homo sapiens (Human) | PR |
| Q9NX65 | ZSCAN32 | Zinc finger and SCAN domain-containing protein 32 | Homo sapiens (Human) | PR |
| O14771 | ZNF213 | Zinc finger protein 213 | Homo sapiens (Human) | PR |
| Q8IWY8 | ZSCAN29 | Zinc finger and SCAN domain-containing protein 29 | Homo sapiens (Human) | PR |
| Q8NCP5 | ZBTB44 | Zinc finger and BTB domain-containing protein 44 | Homo sapiens (Human) | PR |
| P41182 | BCL6 | B-cell lymphoma 6 protein | Homo sapiens (Human) | PR |
| Q9NQX0 | PRDM6 | Putative histone-lysine N-methyltransferase PRDM6 | Homo sapiens (Human) | PR |
| Q9BU19 | ZNF692 | Zinc finger protein 692 | Homo sapiens (Human) | PR |
| Q08AG5 | ZNF844 | Zinc finger protein 844 | Homo sapiens (Human) | PR |
| Q6R2W3 | ZBED9 | SCAN domain-containing protein 3 | Homo sapiens (Human) | PR |
| P98182 | ZNF200 | Zinc finger protein 200 | Homo sapiens (Human) | PR |
| Q9UK11 | ZNF223 | Zinc finger protein 223 | Homo sapiens (Human) | PR |
| O15156 | ZBTB7B | Zinc finger and BTB domain-containing protein 7B | Homo sapiens (Human) | PR |
| Q6ZMS7 | ZNF783 | Zinc finger protein 783 | Homo sapiens (Human) | PR |
| P59923 | ZNF445 | Zinc finger protein 445 | Homo sapiens (Human) | PR |
| Q8N859 | ZNF713 | Zinc finger protein 713 | Homo sapiens (Human) | PR |
| Q99612 | KLF6 | Krueppel-like factor 6 | Homo sapiens (Human) | PR |
| Q8TD17 | ZNF398 | Zinc finger protein 398 | Homo sapiens (Human) | PR |
| P52739 | ZNF131 | Zinc finger protein 131 | Homo sapiens (Human) | PR |
| A6NGD5 | ZSCAN5C | Zinc finger and SCAN domain-containing protein 5C | Homo sapiens (Human) | PR |
| Q05215 | EGR4 | Early growth response protein 4 | Homo sapiens (Human) | PR |
| Q7Z398 | ZNF550 | Zinc finger protein 550 | Homo sapiens (Human) | PR |
| Q9Y2K1 | ZBTB1 | Zinc finger and BTB domain-containing protein 1 | Homo sapiens (Human) | PR |
| Q96N20 | ZNF75A | Zinc finger protein 75A | Homo sapiens (Human) | PR |
| A6NJL1 | ZSCAN5B | Zinc finger and SCAN domain-containing protein 5B | Homo sapiens (Human) | PR |
| A1YPR0 | ZBTB7C | Zinc finger and BTB domain-containing protein 7C | Homo sapiens (Human) | PR |
| Q9NWS9 | ZNF446 | Zinc finger protein 446 | Homo sapiens (Human) | PR |
| P24278 | ZBTB25 | Zinc finger and BTB domain-containing protein 25 | Homo sapiens (Human) | PR |
| Q96N38 | ZNF714 | Zinc finger protein 714 | Homo sapiens (Human) | PR |
| Q86YH2 | ZNF280B | Zinc finger protein 280B | Homo sapiens (Human) | PR |
| O08584 | Klf6 | Krueppel-like factor 6 | Mus musculus (Mouse) | PR |
| Q61164 | Ctcf | Transcriptional repressor CTCF | Mus musculus (Mouse) | PR |
| Q810A1 | Znf18 | Zinc finger protein 18 | Mus musculus (Mouse) | PR |
| Q8BGS3 | Zkscan1 | Zinc finger protein with KRAB and SCAN domains 1 | Mus musculus (Mouse) | PR |
| Q00899 | Yy1 | Transcriptional repressor protein YY1 | Mus musculus (Mouse) | PR |
| P41183 | Bcl6 | B-cell lymphoma 6 protein homolog | Mus musculus (Mouse) | PR |
| Q9DAI4 | Zbtb43 | Zinc finger and BTB domain-containing protein 43 | Mus musculus (Mouse) | PR |
| O70237 | Gfi1b | Zinc finger protein Gfi-1b | Mus musculus (Mouse) | PR |
| Q99KZ6 | Znf639 | Zinc finger protein 639 | Mus musculus (Mouse) | PR |
| Q9Z1D9 | Znf394 | Zinc finger protein 394 | Mus musculus (Mouse) | PR |
| Q9CXE0 | Prdm5 | PR domain zinc finger protein 5 | Mus musculus (Mouse) | PR |
| P43300 | Egr3 | Early growth response protein 3 | Mus musculus (Mouse) | PR |
| Q9DAU9 | Znf654 | Zinc finger protein 654 | Mus musculus (Mouse) | PR |
| Q9R1Y5 | Hic1 | Hypermethylated in cancer 1 protein | Mus musculus (Mouse) | PR |
| Q8R0T2 | Znf768 | Zinc finger protein 768 | Mus musculus (Mouse) | PR |
| Q8BI73 | Znf775 | Zinc finger protein 775 | Mus musculus (Mouse) | PR |
| Q8VCZ7 | Zbtb7c | Zinc finger and BTB domain-containing protein 7C | Mus musculus (Mouse) | PR |
| Q91VN1 | Znf24 | Zinc finger protein 24 | Mus musculus (Mouse) | PR |
| Q9DB38 | Znf580 | Zinc finger protein 580 | Mus musculus (Mouse) | PR |
| A7KBS4 | Zscan4d | Zinc finger and SCAN domain containing protein 4D | Mus musculus (Mouse) | PR |
| Q91VW9 | Zkscan3 | Zinc finger protein with KRAB and SCAN domains 3 | Mus musculus (Mouse) | PR |
| P10925 | Zfy1 | Zinc finger Y-chromosomal protein 1 | Mus musculus (Mouse) | PR |
| P08046 | Egr1 | Early growth response protein 1 | Mus musculus (Mouse) | PR |
| Q3TTC2 | Yy2 | Transcription factor YY2 | Mus musculus (Mouse) | PR |
| Q3UTQ7 | Prdm10 | PR domain zinc finger protein 10 | Mus musculus (Mouse) | PR |
| Q6P3Y5 | Znf280c | Zinc finger protein 280C | Mus musculus (Mouse) | PR |
| Q9ERU3 | Znf22 | Zinc finger protein 22 | Mus musculus (Mouse) | PR |
| Q8VIG1 | Rest | RE1-silencing transcription factor | Mus musculus (Mouse) | PR |
| Q9Z1D8 | Zkscan5 | Zinc finger protein with KRAB and SCAN domains 5 | Mus musculus (Mouse) | PR |
| Q8BID6 | Zbtb46 | Zinc finger and BTB domain-containing protein 46 | Mus musculus (Mouse) | PR |
| P17012 | Zfx | Zinc finger X-chromosomal protein | Mus musculus (Mouse) | PR |
| Q9WUK6 | Zbtb18 | Zinc finger and BTB domain-containing protein 18 | Mus musculus (Mouse) | PR |
| O35738 | Klf12 | Krueppel-like factor 12 | Mus musculus (Mouse) | PR |
| B2RXC5 | Znf382 | Zinc finger protein 382 | Mus musculus (Mouse) | PR |
| O08900 | Ikzf3 | Zinc finger protein Aiolos | Mus musculus (Mouse) | PR |
| Q5DU09 | Znf652 | Zinc finger protein 652 | Mus musculus (Mouse) | PR |
| Q5RJ54 | Zscan26 | Zinc finger and SCAN domain-containing protein 26 | Mus musculus (Mouse) | PR |
| Q8BLM0 | Klf8 | Krueppel-like factor 8 | Mus musculus (Mouse) | PR |
| Q99JB0 | Klf7 | Krueppel-like factor 7 | Mus musculus (Mouse) | PR |
| Q8R0A2 | Zbtb44 | Zinc finger and BTB domain-containing protein 44 | Mus musculus (Mouse) | PR |
| P20662 | Zfy2 | Zinc finger Y-chromosomal protein 2 | Mus musculus (Mouse) | PR |
| Q80VJ6 | Zscan4c | Zinc finger and SCAN domain containing protein 4C | Mus musculus (Mouse) | PR |
| Q3URS2 | Zscan4f | Zinc finger and SCAN domain containing protein 4F | Mus musculus (Mouse) | PR |
| Q60980 | Klf3 | Krueppel-like factor 3 | Mus musculus (Mouse) | PR |
| Q8K3J5 | Znf131 | Zinc finger protein 131 | Mus musculus (Mouse) | PR |
| Q9WVG7 | Osr1 | Protein odd-skipped-related 1 | Mus musculus (Mouse) | PR |
| Q9Z2K3 | Znf394 | Zinc finger protein 394 | Rattus norvegicus (Rat) | PR |
| Q642B9 | Znf18 | Zinc finger protein 18 | Rattus norvegicus (Rat) | PR |
| D3ZUU2 | Gzf1 | GDNF-inducible zinc finger protein 1 | Rattus norvegicus (Rat) | PR |
| B1WBU4 | Zbtb8a | Zinc finger and BTB domain-containing protein 8A | Rattus norvegicus (Rat) | PR |
| Q7TNK3 | Znf24 | Zinc finger protein 24 | Rattus norvegicus (Rat) | PR |
| O35819 | Klf6 | Krueppel-like factor 6 | Rattus norvegicus (Rat) | PR |
| Q9R1D1 | Ctcf | Transcriptional repressor CTCF | Rattus norvegicus (Rat) | PR |
| P43301 | Egr3 | Early growth response protein 3 | Rattus norvegicus (Rat) | PR |
| P08154 | Egr1 | Early growth response protein 1 | Rattus norvegicus (Rat) | PR |
| A0JPL0 | Znf382 | Zinc finger protein 382 | Rattus norvegicus (Rat) | PR |
| Q4KLI1 | Zkscan1 | Zinc finger protein with KRAB and SCAN domains 1 | Rattus norvegicus (Rat) | PR |
| A1L1J6 | Znf652 | Zinc finger protein 652 | Rattus norvegicus (Rat) | PR |
| B0K011 | Osr1 | Protein odd-skipped-related 1 | Rattus norvegicus (Rat) | PR |
| Q9SHD0 | ZAT4 | Zinc finger protein ZAT4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0P4X6 | zbtb44 | Zinc finger and BTB domain-containing protein 44 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| A4II20 | egr1 | Early growth response protein 1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q6P882 | zbtb8a.2 | Zinc finger and BTB domain-containing protein 8A.2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q567C6 | znf367 | Zinc finger protein 367 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| A7Y7X5 | znf711 | Zinc finger protein 711 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGSKTLPAPV | PIHPSLQLTN | YSFLQAVNGL | PTVPSDHLPN | LYGFSALHAV | HLHQWTLGYP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AMHLPRSSFS | KVPGTVSSLV | DARFQLPAFP | WFPHVIQPKP | EITAGGSVPA | LKTKPRFDFA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NLALAATQED | PAKLGRGEGP | GSPAGGLGAL | LDVTKLSPEK | KPTRGRLPSK | TKKEFVCKFC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GRHFTKSYNL | LIHERTHTDE | RPYTCDICHK | AFRRQDHLRD | HRYIHSKEKP | FKCQECGKGF |
| 250 | 260 | ||||
| CQSRTLAVHK | TLHSQVKELK | TSKIKC |