Q8NHV9
Gene name |
RHOXF1 |
Protein name |
Rhox homeobox family member 1 |
Names |
Ovary-, testis- and epididymis-expressed gene protein, Paired-like homeobox protein PEPP-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:158800 |
EC number |
|
Protein Class |
|
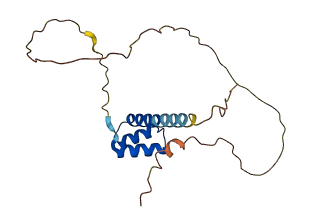
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q8NHV9
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8NHV9-F1 | Predicted | AlphaFoldDB |
141 variants for Q8NHV9
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA414390817 rs782532336 |
2 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782532336 CA10504181 |
2 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782627066 CA10504179 |
3 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA414390814 rs1556000516 |
3 | R>H | No |
ClinGen gnomAD |
|
|
CA414390806 rs1556000513 |
4 | S>L | No |
ClinGen gnomAD |
|
|
CA414390811 rs1556000514 |
4 | S>T | No |
ClinGen gnomAD |
|
|
CA414390805 rs1407807674 |
5 | L>I | No |
ClinGen TOPMed |
|
|
rs1556000507 CA414390796 |
6 | V>A | No |
ClinGen gnomAD |
|
|
CA414390798 rs931167506 |
6 | V>F | No |
ClinGen TOPMed gnomAD |
|
|
CA414390799 rs931167506 |
6 | V>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA334995577 rs931167506 |
6 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
rs781806029 CA414390793 |
7 | H>D | No |
ClinGen ExAC gnomAD |
|
|
rs1240481224 CA414390791 |
7 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
rs781806029 CA10504177 |
7 | H>Y | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 8 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10504176 COSM1223720 rs782699512 |
8 | D>N | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA414390779 rs1556000500 |
9 | T>A | No |
ClinGen gnomAD |
|
|
rs149847534 CA10504174 |
10 | V>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA414390774 rs868930445 |
10 | V>M | No |
ClinGen gnomAD |
|
|
CA10504173 rs782766172 |
11 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA10504170 rs202119409 |
15 | S>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs782043508 CA10504168 |
21 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA414390655 rs782412199 |
27 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782412199 CA10504167 |
27 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA334995575 rs928542538 |
28 | G>A | No |
ClinGen Ensembl |
|
|
CA414390653 rs782270842 |
28 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504166 rs782270842 |
28 | G>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs980162869 CA334995574 |
29 | A>V | No |
ClinGen Ensembl |
|
|
rs781990338 CA10504165 |
30 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782356480 CA10504164 |
31 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA414390629 rs1377162159 |
32 | S>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA414390627 rs1435468418 |
33 | A>T | No |
ClinGen TOPMed |
|
|
CA10504162 rs782576838 |
36 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA10504160 rs36014613 |
37 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1556000479 CA414390572 |
41 | A>D | No |
ClinGen gnomAD |
|
|
CA10504158 rs782513988 |
44 | L>V | No |
ClinGen ExAC |
|
|
rs781846970 CA10504157 |
45 | M>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 47 | N>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10504155 rs782456853 |
48 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA10504154 rs140029775 |
50 | P>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA334995573 rs990207644 |
51 | E>* | No |
ClinGen Ensembl |
|
|
CA10504152 rs782155047 |
52 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782155047 CA414390499 |
52 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs150693893 CA10504153 COSM3964633 |
52 | G>S | lung [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1556000472 CA414390496 |
53 | G>C | No |
ClinGen gnomAD |
|
|
rs1556000470 CA414390485 |
55 | N>D | No |
ClinGen gnomAD |
|
|
rs1556000468 CA414390482 |
55 | N>S | No |
ClinGen gnomAD |
|
|
CA10504150 rs782764697 |
57 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA10504149 rs782101893 |
57 | E>V | No |
ClinGen ExAC gnomAD |
|
|
CA10504147 rs782032966 |
59 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414390455 rs868942951 COSM247038 |
59 | G>D | Variant assessed as Somatic; impact. prostate [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
|
rs782032966 CA10504146 |
59 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782032966 CA10504145 |
59 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA334995572 rs868942951 |
59 | G>V | No |
ClinGen TOPMed |
|
|
rs1339647526 CA414390433 |
62 | R>P | No |
ClinGen TOPMed |
|
|
CA414390430 rs1556000460 |
63 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs782630665 CA10504142 |
64 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA10504141 rs782482911 |
65 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414390416 COSM755632 rs868960412 |
65 | G>D | lung large_intestine [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA10504140 rs782202572 |
66 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1556000455 CA414390404 |
67 | I>V | No |
ClinGen gnomAD |
|
|
rs781893705 CA414390387 |
69 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504138 rs782569899 |
69 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1454758659 CA414390379 |
71 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
rs782783073 CA414390372 |
72 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504136 rs782783073 |
72 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781841223 CA414390343 |
76 | E>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781841223 CA10504134 |
76 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414390346 rs1449125054 |
76 | E>K | No |
ClinGen TOPMed |
|
|
CA414390335 rs1556000440 |
77 | P>R | No |
ClinGen gnomAD |
|
|
CA10504132 rs782062680 |
78 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504131 rs782174738 COSM3390392 |
78 | R>Q | Variant assessed as Somatic; 0.0 impact. pancreas [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs782801044 CA10504130 |
80 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1362525858 CA414390306 |
82 | Q>P | No |
ClinGen TOPMed |
|
|
CA414390305 rs1362525858 |
82 | Q>R | No |
ClinGen TOPMed |
|
|
CA414390294 rs781993646 |
84 | P>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs781996156 CA10504127 |
84 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs781996156 CA414390292 |
84 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs781993646 CA10504128 |
84 | P>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA10504125 rs782774382 |
85 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA414390272 rs1309600311 |
87 | E>D | No |
ClinGen TOPMed |
|
|
CA10504123 rs782318799 |
88 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA414390267 rs782318799 |
88 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504124 rs782318799 |
88 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504122 rs782298395 |
89 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414390259 rs1556000405 |
90 | Q>* | No |
ClinGen gnomAD |
|
|
CA414390250 rs1246288348 |
91 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
| TCGA novel | 92 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA414390237 rs1556000400 |
93 | M>T | No |
ClinGen gnomAD |
|
|
rs880000723 CA414390240 |
93 | M>V | No |
ClinGen gnomAD |
|
|
CA414390221 rs782682773 |
95 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504120 rs782390666 |
95 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782682773 CA10504121 |
95 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782248993 CA10504119 |
96 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA414390199 rs1416988640 |
99 | E>K | No |
ClinGen TOPMed |
|
|
CA414390179 rs1556000395 |
101 | M>K | No |
ClinGen gnomAD |
|
|
CA10504117 rs368824826 |
101 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10504116 rs781960574 |
103 | P>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA414390159 rs1556000394 |
104 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs782571468 CA10504115 |
105 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA334995568 rs781931824 |
106 | R>P | No |
ClinGen TOPMed gnomAD |
|
|
COSM1465244 rs1385779747 CA414390139 |
108 | T>M | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA TOPMed |
| TCGA novel | 115 | V>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs938508848 CA334995565 |
118 | L>M | No |
ClinGen TOPMed |
|
|
CA10504113 rs781885702 |
120 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1293096269 CA414390058 |
120 | S>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA414390039 rs1336042822 |
123 | R>G | No |
ClinGen TOPMed |
|
|
rs782789749 CA10504112 |
123 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA414390018 rs1556000379 |
126 | Q>P | No |
ClinGen gnomAD |
|
|
rs1556000378 CA414390006 |
127 | Y>* | No |
ClinGen Ensembl |
|
|
CA10504110 rs781837641 |
131 | P>H | No |
ClinGen ExAC gnomAD |
|
|
CA10504108 rs374507906 |
132 | T>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs374507906 CA10504109 |
132 | T>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 137 | A>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 138 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10504089 rs191816066 |
138 | E>K | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA414389522 rs1426587023 |
139 | N>T | No |
ClinGen TOPMed |
|
|
CA414389470 rs1569312180 |
144 | E>K | No |
ClinGen Ensembl |
|
|
rs782781485 CA334995416 |
148 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs782781485 CA10504088 |
148 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs147476615 CA10504074 |
158 | C>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10504073 rs782429266 |
158 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1223192137 CA414389016 |
160 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1484612375 CA414388930 |
168 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1204114561 CA414388923 |
168 | A>V | No |
ClinGen TOPMed |
|
|
CA10504071 rs148295665 |
169 | N>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs144256540 CA10504070 |
170 | E>V | No |
ClinGen ESP ExAC TOPMed |
|
|
rs781847765 CA10504069 |
171 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA414388887 rs1555999424 |
172 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs2301977 VAR_049587 RCV000888556 CA10504068 |
172 | R>H | found in infertile men; unknown pathological significance; no effect on induction of target genes expression [UniProt] | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA414388869 rs1445759218 |
174 | D>Y | No |
ClinGen TOPMed |
|
|
CA414388856 rs1555999416 |
175 | P>S | No |
ClinGen gnomAD |
|
|
CA10504063 rs373236335 |
176 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504065 rs782819273 |
176 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA334995208 rs782813436 |
177 | D>G | No |
ClinGen gnomAD |
|
|
VAR_077002 CA10504062 rs138060880 |
177 | D>H | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA10504061 rs138060880 |
177 | D>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1555999399 CA414388794 |
181 | I>V | No |
ClinGen gnomAD |
|
|
rs782317805 CA10504059 |
182 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10504057 rs782024712 |
183 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA10504056 rs782395381 |
184 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
No associated diseases with Q8NHV9
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| androgen receptor signaling pathway | The series of molecular signals initiated by androgen binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| enteric nervous system development | The process whose specific outcome is the progression of the enteric nervous system over time, from its formation to the mature structure. The enteric nervous system is composed of two ganglionated neural plexuses in the gut wall which form one of the three major divisions of the autonomic nervous system. The enteric nervous system innervates the gastrointestinal tract, the pancreas, and the gall bladder. It contains sensory neurons, interneurons, and motor neurons. Thus the circuitry can autonomously sense the tension and the chemical environment in the gut and regulate blood vessel tone, motility, secretions, and fluid transport. The system is itself governed by the central nervous system and receives both parasympathetic and sympathetic innervation. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| regulation of DNA-templated transcription | Any process that modulates the frequency, rate or extent of cellular DNA-templated transcription. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
22 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A0JNI8 | LHX9 | LIM/homeobox protein Lhx9 | Bos taurus (Bovine) | PR |
| Q90881 | LHX9 | LIM/homeobox protein Lhx9 | Gallus gallus (Chicken) | PR |
| F1NEA7 | DMBX1 | Diencephalon/mesencephalon homeobox protein 1 | Gallus gallus (Chicken) | PR |
| Q90963 | PRRX2 | Paired mesoderm homeobox protein 2 | Gallus gallus (Chicken) | PR |
| Q8IRC7 | Awh | LIM/homeobox protein Awh | Drosophila melanogaster (Fruit fly) | PR |
| Q9NQ69 | LHX9 | LIM/homeobox protein Lhx9 | Homo sapiens (Human) | PR |
| Q99811 | PRRX2 | Paired mesoderm homeobox protein 2 | Homo sapiens (Human) | PR |
| P50458 | LHX2 | LIM/homeobox protein Lhx2 | Homo sapiens (Human) | PR |
| O15266 | SHOX | Short stature homeobox protein | Homo sapiens (Human) | PR |
| O95076 | ALX3 | Homeobox protein aristaless-like 3 | Homo sapiens (Human) | PR |
| Q9BQY4 | RHOXF2 | Rhox homeobox family member 2 | Homo sapiens (Human) | PR |
| O14813 | PHOX2A | Paired mesoderm homeobox protein 2A | Homo sapiens (Human) | PR |
| P63013 | Prrx1 | Paired mesoderm homeobox protein 1 | Mus musculus (Mouse) | PR |
| Q8VIH1 | Nobox | Homeobox protein NOBOX | Mus musculus (Mouse) | PR |
| Q06348 | Prrx2 | Paired mesoderm homeobox protein 2 | Mus musculus (Mouse) | PR |
| O88933 | Esx1 | Extraembryonic, spermatogenesis, homeobox 1 (Homeobox protein SPX1) (Homeodomain protein EPX) | Mus musculus (Mouse) | EV |
| Q9WUH2 | Lhx9 | LIM/homeobox protein Lhx9 | Mus musculus (Mouse) | PR |
| Q80W90 | Lhx9 | LIM/homeobox protein Lhx9 | Rattus norvegicus (Rat) | PR |
| P36198 | Lhx2 | LIM/homeobox protein Lhx2 | Rattus norvegicus (Rat) | PR |
| Q28G02 | siamois | Homeobox protein siamois | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q566X8 | dmbx1b | Diencephalon/mesencephalon homeobox protein 1-B | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q1LWV4 | lhx9 | LIM/homeobox protein Lhx9 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MARSLVHDTV | FYCLSVYQVK | ISPTPQLGAA | SSAEGHVGQG | APGLMGNMNP | EGGVNHENGM |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NRDGGMIPEG | GGGNQEPRQQ | PQPPPEEPAQ | AAMEGPQPEN | MQPRTRRTKF | TLLQVEELES |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VFRHTQYPDV | PTRRELAENL | GVTEDKVRVW | FKNKRARCRR | HQRELMLANE | LRADPDDCVY |
| IVVD |