Q8NFU1
Gene name |
BEST2 (VMD2L1) |
Protein name |
Bestrophin-2a |
Names |
Vitelliform macular dystrophy 2-like protein 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:54831 |
EC number |
|
Protein Class |
|
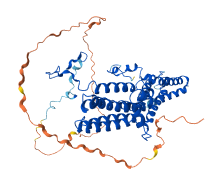
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1-421 (Bestrophin) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for Q8NFU1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 8D1E | EM | 178 A | A/B/C/D/E | 1-406 | PDB |
| 8D1F | EM | 182 A | A/B/C/D/E | 1-406 | PDB |
| 8D1G | EM | 207 A | A/B/C/D/E | 1-406 | PDB |
| 8D1H | EM | 194 A | A/B/C/D/E | 1-406 | PDB |
| AF-Q8NFU1-F1 | Predicted | AlphaFoldDB |
605 variants for Q8NFU1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs762982612 | 3 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs766284816 | 4 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1967881949 | 5 | Y>* | No | TOPMed | |
| rs185747118 | 7 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs79300835 | 8 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs79300835 | 8 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1568351857 | 9 | V>E | No | Ensembl | |
| rs1967882322 | 9 | V>L | No |
TOPMed gnomAD |
|
| rs755678866 | 10 | A>V | No |
ExAC gnomAD |
|
| rs750931656 | 11 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1320783177 | 11 | N>T | No | gnomAD | |
| rs780490794 | 12 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs560892056 | 12 | A>V | No |
TOPMed gnomAD |
|
| rs747298154 | 13 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs747298154 | 13 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs368197840 | 13 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs368197840 | 13 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs747298154 | 13 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1967883300 | 15 | G>D | No | Ensembl | |
| rs374373357 | 15 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs374373357 | 15 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs770068797 | 17 | F>Y | No |
ExAC gnomAD |
|
| rs1568351890 | 18 | S>F | No | Ensembl | |
| rs773361477 | 19 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs1967883738 | 19 | Q>R | No | TOPMed | |
| rs1324448850 | 21 | L>P | No | gnomAD | |
| TCGA novel | 23 | L>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1487952926 | 23 | L>P | No | gnomAD | |
| rs368305126 | 25 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs770731972 | 25 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1245830380 | 26 | G>R | No | TOPMed | |
| rs1245830380 | 26 | G>W | No | TOPMed | |
| rs1967884300 | 28 | I>V | No | Ensembl | |
| COSM3796648 | 30 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775071591 | 33 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs760251223 | 34 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs763605475 | 34 | R>P | No |
ExAC TOPMed gnomAD |
|
|
COSM159365 rs763605475 |
34 | R>Q | breast [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1419675927 | 35 | E>* | No | gnomAD | |
| rs1313838971 | 37 | L>P | No | gnomAD | |
| rs371908216 | 38 | C>S | No |
ESP ExAC gnomAD |
|
| rs1394680086 | 39 | F>L | No | gnomAD | |
| TCGA novel | 42 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369765091 | 43 | Y>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1340233902 | 44 | M>L | No |
TOPMed gnomAD |
|
| rs1306514244 | 44 | M>T | No | gnomAD | |
| rs190553647 | 45 | A>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1297172840 | 45 | A>T | No | TOPMed | |
| rs190553647 | 45 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1342541998 | 47 | S>T | No | gnomAD | |
| rs1967885486 | 48 | A>G | No | Ensembl | |
| rs1967885486 | 48 | A>V | No | Ensembl | |
| rs756350492 | 51 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs376542599 | 51 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1365379560 | 53 | V>M | No | gnomAD | |
| rs779857048 | 54 | L>P | No |
ExAC gnomAD |
|
|
rs768418645 COSM438624 |
56 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs768418645 | 56 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs552068493 | 57 | G>A | No | gnomAD | |
| COSM2151828 | 58 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1568352114 | 59 | K>R | No | gnomAD | |
| rs1568352114 | 59 | K>T | No | gnomAD | |
| rs201824648 | 60 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs200640940 COSM3378615 |
60 | R>H | pancreas [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs772723089 | 62 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs769196924 | 62 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs571724788 | 63 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs932982100 | 64 | K>Q | No | Ensembl | |
| rs376438832 | 65 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs376438832 | 65 | L>R | No |
ESP ExAC TOPMed gnomAD |
|
| COSM6149596 | 69 | C>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780644746 | 69 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs1347572596 | 70 | D>Y | No | gnomAD | |
| rs1221482096 | 71 | Q>K | No | gnomAD | |
| COSM4074439 | 71 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs371008787 | 72 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs778880279 | 77 | P>L | No |
ExAC gnomAD |
|
| TCGA novel | 77 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1297419517 | 78 | V>A | No | gnomAD | |
| rs750474711 | 78 | V>I | No |
ExAC TOPMed gnomAD |
|
|
rs750474711 COSM1304039 |
78 | V>L | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1340101300 | 79 | S>P | No | gnomAD | |
| rs1340101300 | 79 | S>T | No | gnomAD | |
| TCGA novel | 79 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201108396 | 80 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1234084795 | 80 | F>S | No | gnomAD | |
| rs746437107 | 81 | V>L | No |
TOPMed gnomAD |
|
| rs746437107 | 81 | V>M | No |
TOPMed gnomAD |
|
| rs1418231183 | 82 | L>F | No | gnomAD | |
| rs1967894975 | 83 | G>C | No |
TOPMed gnomAD |
|
| rs867186836 | 84 | F>L | No | Ensembl | |
| rs1430181591 | 85 | Y>* | No | TOPMed | |
| rs944968547 | 86 | V>A | No | Ensembl | |
| rs751506190 | 87 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1039150174 | 87 | T>S | No | Ensembl | |
| rs1967912571 | 88 | L>V | No | TOPMed | |
| rs1244925823 | 91 | N>D | No | gnomAD | |
| rs1204597745 | 91 | N>K | No |
TOPMed gnomAD |
|
| rs1599457652 | 91 | N>T | No | Ensembl | |
| rs1379146201 | 92 | R>C | No | gnomAD | |
| rs754854813 | 92 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1372449301 | 93 | W>* | No | gnomAD | |
| rs1277012963 | 95 | S>R | No |
TOPMed gnomAD |
|
| rs1311222576 | 96 | Q>* | No | gnomAD | |
| rs201615329 | 97 | Y>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201615329 | 97 | Y>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs368638952 | 98 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1478995100 | 100 | M>I | No | gnomAD | |
| rs1209078342 | 100 | M>K | No | Ensembl | |
| rs1347434462 | 101 | P>L | No | TOPMed | |
| rs868198303 | 101 | P>S | No | Ensembl | |
| rs1424155955 | 103 | P>S | No | gnomAD | |
| rs748886255 | 104 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs777534813 | 104 | D>G | No |
ExAC gnomAD |
|
| rs1967913437 | 104 | D>H | No |
TOPMed gnomAD |
|
| rs1599457699 | 105 | A>T | No | Ensembl | |
| rs770565791 | 105 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1273618050 | 107 | M>I | No | gnomAD | |
| rs773939596 | 107 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1367266873 | 108 | C>F | No |
TOPMed gnomAD |
|
| rs372694212 | 108 | C>R | No |
ESP TOPMed |
|
| rs769244875 | 109 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1315753554 | 110 | V>L | No |
1000Genomes TOPMed gnomAD |
|
| rs1967913914 | 111 | A>V | No | Ensembl | |
| rs936540167 | 112 | G>S | No |
TOPMed gnomAD |
|
| rs1967914036 | 113 | T>A | No | TOPMed | |
| rs762288691 | 113 | T>N | No |
ExAC gnomAD |
|
| rs375044336 | 114 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs375044336 | 114 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs773552289 | 115 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs1967914199 | 115 | H>R | No | gnomAD | |
| rs1475585144 | 116 | G>* | No |
TOPMed gnomAD |
|
| rs1475585144 | 116 | G>R | No |
TOPMed gnomAD |
|
| rs1967914361 | 117 | R>G | No | TOPMed | |
| rs1967914399 | 117 | R>H | No | TOPMed | |
| rs1967914399 | 117 | R>L | No | TOPMed | |
| rs1463366835 | 118 | D>H | No | TOPMed | |
| rs1463366835 | 118 | D>N | No | TOPMed | |
| rs763096626 | 119 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1476986043 | 120 | R>C | No |
TOPMed gnomAD |
|
| rs561379374 | 120 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1476986043 | 120 | R>S | No |
TOPMed gnomAD |
|
| rs1407181245 | 121 | G>A | No | gnomAD | |
| rs751592629 | 121 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1407181245 | 121 | G>D | No | gnomAD | |
| rs751592629 | 121 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1407181245 | 121 | G>V | No | gnomAD | |
| rs1322087639 | 123 | L>H | No |
TOPMed gnomAD |
|
| rs1050010291 | 125 | R>Q | No |
TOPMed gnomAD |
|
| rs2145704313 | 126 | R>G | No | Ensembl | |
| rs1443670092 | 126 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs767536264 | 128 | L>V | No |
ExAC gnomAD |
|
| rs1284457717 | 129 | M>I | No | gnomAD | |
| rs1357769318 | 130 | R>H | No |
TOPMed gnomAD |
|
| rs1226130360 | 131 | Y>C | No | gnomAD | |
| rs756001142 | 132 | A>S | No |
ExAC gnomAD |
|
| COSM991433 | 132 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1218478174 | 132 | A>V | No | gnomAD | |
| rs1487850524 | 133 | G>E | No | gnomAD | |
| rs1193162226 | 134 | L>P | No | gnomAD | |
| rs371302296 | 135 | S>L | No |
ESP TOPMed gnomAD |
|
| rs1161606512 | 136 | A>D | No |
TOPMed gnomAD |
|
| rs1161606512 | 136 | A>V | No |
TOPMed gnomAD |
|
| rs745409765 | 137 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs745409765 | 137 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs778596981 | 137 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs778596981 | 137 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1967915665 | 138 | L>F | No | Ensembl | |
| rs1358980288 | 138 | L>H | No | gnomAD | |
| rs771687530 | 139 | I>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 139 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs199593030 | 141 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1308306371 | 142 | S>C | No |
TOPMed gnomAD |
|
| rs1967915983 | 143 | V>A | No | gnomAD | |
|
COSM1246085 rs565237088 |
143 | V>I | oesophagus [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs565237088 | 143 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1242165153 | 145 | T>I | No | gnomAD | |
| rs1352621506 | 146 | A>T | No | gnomAD | |
| rs1200886719 | 146 | A>V | No | gnomAD | |
|
rs1185484635 COSM318916 |
150 | R>L | lung [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1185484635 | 150 | R>P | No |
TOPMed gnomAD |
|
| rs1236928473 | 151 | F>L | No | gnomAD | |
|
COSM3528824 rs1162832309 |
152 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs532624266 | 154 | I>V | No | 1000Genomes | |
| rs771105784 | 155 | D>H | No |
ExAC gnomAD |
|
| rs771105784 | 155 | D>N | No |
ExAC gnomAD |
|
| rs1173595749 | 157 | V>M | No |
TOPMed gnomAD |
|
| rs1412449862 | 158 | V>A | No | gnomAD | |
| rs34896167 | 158 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1250178650 | 160 | A>G | No | TOPMed | |
| rs1967918612 | 161 | G>E | No | Ensembl | |
| rs865890676 | 161 | G>R | No | TOPMed | |
| rs865890676 | 161 | G>W | No | TOPMed | |
| rs376890260 | 162 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1599457992 | 162 | F>V | No | Ensembl | |
| rs1967918764 | 163 | M>R | No | Ensembl | |
| rs1295159301 | 165 | R>C | No |
TOPMed gnomAD |
|
|
COSM3959556 rs2145704645 |
165 | R>H | lung [Cosmic] | No |
cosmic curated Ensembl |
| rs1386004275 | 166 | E>D | No | gnomAD | |
| rs922110604 | 166 | E>K | No |
TOPMed gnomAD |
|
| rs1407840110 | 168 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
rs925250751 COSM274131 |
168 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed gnomAD |
| rs925250751 | 168 | R>P | No |
TOPMed gnomAD |
|
| rs1407840110 | 168 | R>S | No |
TOPMed gnomAD |
|
| rs1967919266 | 171 | F>L | No | TOPMed | |
| rs867456508 | 171 | F>S | No | gnomAD | |
| COSM3528826 | 172 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs750107042 | 173 | N>K | No |
ExAC gnomAD |
|
| rs757932379 | 176 | S>A | No |
ExAC gnomAD |
|
| rs1967919476 | 177 | S>P | No | TOPMed | |
| rs750995891 | 179 | N>Y | No |
ExAC gnomAD |
|
| rs754484727 | 181 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs1568352644 | 182 | W>L | No | Ensembl | |
| rs1222192145 | 182 | W>R | No | gnomAD | |
| rs1278632579 | 183 | V>M | No | gnomAD | |
| rs370451899 | 186 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs370451899 | 186 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs202075833 | 187 | W>* | No |
1000Genomes ExAC gnomAD |
|
| rs1190973806 | 188 | F>C | No | gnomAD | |
|
COSM4841097 rs1967919858 |
188 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA Cosmic |
| rs779188520 | 189 | S>A | No |
ExAC gnomAD |
|
| rs1967919999 | 189 | S>Y | No | Ensembl | |
| rs1967920049 | 190 | N>S | No | TOPMed | |
| rs746121828 | 191 | L>R | No |
ExAC gnomAD |
|
| rs1967920155 | 191 | L>V | No | TOPMed | |
| rs1423990908 | 192 | A>E | No |
TOPMed gnomAD |
|
| rs1366613779 | 192 | A>P | No | gnomAD | |
| rs1366613779 | 192 | A>T | No | gnomAD | |
| rs1423990908 | 192 | A>V | No |
TOPMed gnomAD |
|
| rs1362172300 | 193 | A>V | No | gnomAD | |
| rs747106181 | 194 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs1296513269 | 195 | A>P | No | gnomAD | |
| rs776776362 | 196 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs776776362 | 196 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs761777165 | 196 | R>P | No |
ExAC gnomAD |
|
| rs761777165 | 196 | R>Q | No |
ExAC gnomAD |
|
| rs773094930 | 197 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs773094930 | 197 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs762699876 | 197 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs762699876 | 197 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs762699876 | 197 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs2145704775 | 198 | E>A | No | Ensembl | |
| rs765991431 | 198 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs765991431 | 198 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs751206150 | 199 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs377252718 | 200 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs766942702 | 200 | R>H | No |
ExAC gnomAD |
|
| rs766942702 | 200 | R>L | No |
ExAC gnomAD |
|
| rs1230212102 | 201 | I>T | No | gnomAD | |
| rs1253399633 | 202 | R>G | No |
TOPMed gnomAD |
|
| rs1443513308 | 202 | R>H | No |
TOPMed gnomAD |
|
| rs1253399633 | 202 | R>S | No |
TOPMed gnomAD |
|
| rs757710716 | 203 | D>N | No |
ExAC gnomAD |
|
| rs17706412 | 204 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs967907804 | 204 | N>S | No |
TOPMed gnomAD |
|
| rs746257700 | 205 | S>G | No |
ExAC gnomAD |
|
| rs758767858 | 205 | S>R | No |
ExAC gnomAD |
|
| rs780198148 | 206 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1967922077 | 209 | L>P | No | Ensembl | |
| rs747250749 | 210 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs1967922212 | 210 | L>R | No | TOPMed | |
| rs1967922478 | 212 | E>A | No | Ensembl | |
| rs1310613709 | 212 | E>D | No | gnomAD | |
| rs1452026355 | 212 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1259379657 | 213 | E>G | No |
TOPMed gnomAD |
|
| rs868352030 | 213 | E>K | No | Ensembl | |
| rs758756247 | 216 | V>A | No |
ExAC gnomAD |
|
| rs1489333634 | 218 | R>P | No |
TOPMed gnomAD |
|
|
rs1489333634 COSM1318374 |
218 | R>Q | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs780398879 | 218 | R>W | No |
ExAC gnomAD |
|
| rs952366736 | 219 | G>D | No | Ensembl | |
| rs375581807 | 222 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs781454565 | 223 | M>I | No |
ExAC gnomAD |
|
| rs1318993923 | 224 | L>P | No |
TOPMed gnomAD |
|
| TCGA novel | 226 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769975712 | 226 | H>Y | No | ExAC | |
| rs1408280417 | 227 | Y>* | No |
TOPMed gnomAD |
|
| rs895379664 | 227 | Y>C | No |
TOPMed gnomAD |
|
| COSM991434 | 228 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1328701213 | 229 | W>R | No | gnomAD | |
| rs1351031453 | 230 | I>N | No |
TOPMed gnomAD |
|
| rs1351031453 | 230 | I>T | No |
TOPMed gnomAD |
|
| rs749247851 | 232 | V>A | No |
ExAC gnomAD |
|
|
COSM4074440 rs371801091 |
232 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1967929766 | 233 | P>H | No | TOPMed | |
| rs1967929766 | 233 | P>L | No | TOPMed | |
| rs1439417774 | 234 | L>I | No | TOPMed | |
| rs770640416 | 235 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs774143947 | 236 | Y>* | No |
ExAC gnomAD |
|
| rs759214860 | 237 | T>A | No |
ExAC TOPMed gnomAD |
|
|
COSM4074441 rs866786611 |
237 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs866786611 | 237 | T>R | No |
TOPMed gnomAD |
|
|
COSM78881 rs1568352894 |
238 | Q>* | Variant assessed as Somatic; HIGH impact. ovary large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| COSM991435 | 241 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1314274899 | 241 | T>I | No |
TOPMed gnomAD |
|
| rs766951863 | 242 | I>F | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 242 | I>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1349713103 | 243 | A>S | No |
TOPMed gnomAD |
|
| rs1349713103 | 243 | A>T | No |
TOPMed gnomAD |
|
| rs1249920846 | 243 | A>V | No | Ensembl | |
| rs760039438 | 244 | L>Q | No |
ExAC gnomAD |
|
| rs752977310 | 245 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1243038770 | 246 | S>I | No | gnomAD | |
| rs1440902072 | 248 | F>S | No | gnomAD | |
| rs376124946 | 250 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| COSM4074442 | 250 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2145705426 | 251 | C>F | No | Ensembl | |
| rs1967934086 | 253 | I>F | No | Ensembl | |
| rs1967934145 | 253 | I>T | No | TOPMed | |
| rs1967934086 | 253 | I>V | No | Ensembl | |
| rs757260490 | 255 | R>C | No |
ExAC gnomAD |
|
| rs780573011 | 255 | R>H | No |
TOPMed gnomAD |
|
| rs780573011 | 255 | R>L | No |
TOPMed gnomAD |
|
| rs370748557 | 258 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs370748557 | 258 | L>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs768315666 | 259 | D>E | No |
ExAC gnomAD |
|
| rs746686694 | 259 | D>G | No |
ExAC gnomAD |
|
| COSM1390646 | 259 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749711007 | 260 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1475549309 | 260 | P>S | No |
TOPMed gnomAD |
|
| rs1568353029 | 263 | G>V | No | Ensembl | |
| rs755421562 | 266 | D>E | No | Ensembl | |
| rs1568353035 | 266 | D>Y | No | Ensembl | |
| rs775068118 | 268 | D>G | No |
ExAC gnomAD |
|
|
COSM3528827 rs1233783533 |
268 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1325974343 | 269 | L>I | No | gnomAD | |
| rs1458297511 | 270 | D>E | No |
TOPMed gnomAD |
|
| rs768047440 | 270 | D>H | No |
ExAC gnomAD |
|
| rs768047440 | 270 | D>N | No |
ExAC gnomAD |
|
| COSM991436 | 271 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs377503824 | 272 | C>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs753951285 | 273 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs753951285 | 273 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs1479278070 | 276 | F>V | No |
TOPMed gnomAD |
|
| TCGA novel | 277 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1967935680 | 279 | L>F | No |
TOPMed gnomAD |
|
| rs750419281 | 280 | Q>H | No |
ExAC TOPMed gnomAD |
|
|
rs150329270 COSM1732248 |
285 | A>T | bone [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs754679351 | 286 | G>S | No |
ExAC gnomAD |
|
| rs368957208 | 287 | W>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs1967936047 | 287 | W>G | No | Ensembl | |
| rs747685994 | 288 | L>F | No |
ExAC gnomAD |
|
| TCGA novel | 288 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1967937383 | 290 | V>I | No | gnomAD | |
| rs780712688 | 292 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1967937564 | 293 | Q>* | No | Ensembl | |
| rs1967937607 | 294 | L>F | No | Ensembl | |
| rs1967937649 | 295 | I>V | No | TOPMed | |
| rs1246113501 | 296 | N>S | No | gnomAD | |
| COSM3528828 | 297 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1294590567 | 297 | P>S | No |
TOPMed gnomAD |
|
| rs951262138 | 299 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1211026101 | 300 | E>Q | No | gnomAD | |
| TCGA novel | 301 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1967937947 | 301 | D>N | No | TOPMed | |
| rs769123082 | 302 | D>N | No |
ExAC gnomAD |
|
| COSM3528829 | 303 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776837626 | 304 | D>E | No |
ExAC gnomAD |
|
| rs1967938332 | 311 | I>N | No | Ensembl | |
| rs773372888 | 312 | D>H | No |
ExAC gnomAD |
|
| rs773372888 | 312 | D>Y | No |
ExAC gnomAD |
|
| rs1194636115 | 313 | R>K | No |
TOPMed gnomAD |
|
|
COSM5462799 rs1456000924 |
315 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
TOPMed gnomAD NCI-TCGA Cosmic |
| rs763032729 | 315 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1456000924 | 315 | F>V | No |
TOPMed gnomAD |
|
| rs753604392 | 317 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs756949309 | 318 | S>Y | No |
ExAC gnomAD |
|
| rs764843771 | 319 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1422488005 | 319 | M>V | No |
TOPMed gnomAD |
|
| rs1025130868 | 320 | L>M | No | Ensembl | |
| rs1260645848 | 321 | A>G | No |
TOPMed gnomAD |
|
| rs749932847 | 321 | A>S | No |
ExAC gnomAD |
|
| rs749932847 | 321 | A>T | No |
ExAC gnomAD |
|
| rs1260645848 | 321 | A>V | No |
TOPMed gnomAD |
|
| TCGA novel | 323 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1967941902 | 324 | E>Q | No | TOPMed | |
| rs1199115547 | 325 | M>I | No |
TOPMed gnomAD |
|
| rs1967941928 | 325 | M>T | No | TOPMed | |
| rs781562291 | 326 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs756535904 | 327 | D>V | No |
ExAC gnomAD |
|
| rs1967942054 | 327 | D>Y | No | Ensembl | |
| rs1568353208 | 329 | L>Q | No | Ensembl | |
| rs1967942241 | 333 | E>Q | No |
TOPMed gnomAD |
|
| rs774588558 | 336 | L>F | No |
ExAC gnomAD |
|
| TCGA novel | 337 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs949339910 | 341 | A>D | No | TOPMed | |
| rs772126783 | 341 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1440428384 | 342 | E>A | No | gnomAD | |
| rs775360080 | 342 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1298012946 | 343 | A>T | No |
TOPMed gnomAD |
|
| rs760679073 | 343 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs764012466 | 344 | R>C | No |
TOPMed gnomAD |
|
| rs764031387 | 344 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs764031387 | 344 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs764818855 | 345 | A>T | No |
ExAC gnomAD |
|
| rs1349200958 | 346 | P>L | No | gnomAD | |
| rs765905849 | 347 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs757848832 | 347 | Y>N | No | ExAC | |
| rs1599458993 | 348 | T>P | No | Ensembl | |
| rs367897325 | 349 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| COSM4074445 | 351 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1267109837 | 353 | F>L | No | gnomAD | |
| rs372450964 | 354 | Q>R | No |
ESP TOPMed |
|
| rs1164987892 | 355 | L>R | No | TOPMed | |
| rs202060767 | 356 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs202060767 | 356 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs757587843 COSM1523916 |
356 | R>W | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1967943442 | 358 | P>A | No | TOPMed | |
| rs1408661833 | 358 | P>L | No | gnomAD | |
| rs1366512566 | 359 | S>F | No |
TOPMed gnomAD |
|
| rs746102764 | 359 | S>T | No |
ExAC gnomAD |
|
| rs1599459020 | 364 | T>P | No | Ensembl | |
| rs199683315 | 368 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1967961033 | 370 | A>G | No | gnomAD | |
| rs1004832185 | 372 | E>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1015771882 | 373 | D>E | No |
TOPMed gnomAD |
|
| rs1967961165 | 373 | D>V | No | Ensembl | |
| rs1336870898 | 375 | Q>* | No |
TOPMed gnomAD |
|
| rs1407684322 | 375 | Q>H | No |
TOPMed gnomAD |
|
| rs1356582621 | 375 | Q>P | No | TOPMed | |
| rs781277334 | 377 | Q>* | No |
ExAC gnomAD |
|
| rs1336066713 | 377 | Q>H | No |
TOPMed gnomAD |
|
| rs1967961358 | 377 | Q>R | No | Ensembl | |
| rs1444186809 | 378 | R>G | No |
TOPMed gnomAD |
|
| rs1444186809 | 378 | R>W | No |
TOPMed gnomAD |
|
| rs1281484721 | 379 | L>V | No |
TOPMed gnomAD |
|
| rs1348859638 | 380 | D>E | No |
TOPMed gnomAD |
|
| rs748110787 | 381 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1263790947 | 381 | G>V | No | gnomAD | |
| rs1967961644 | 382 | L>M | No | Ensembl | |
| rs963175145 | 383 | D>E | No |
TOPMed gnomAD |
|
| rs995275607 | 384 | G>E | No |
TOPMed gnomAD |
|
| rs995275607 | 384 | G>V | No |
TOPMed gnomAD |
|
| rs1042649342 | 385 | P>L | No |
TOPMed gnomAD |
|
| rs1402493916 | 385 | P>S | No |
1000Genomes TOPMed gnomAD |
|
| rs1438752808 | 386 | M>T | No |
TOPMed gnomAD |
|
| rs769532316 | 387 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs2145707503 | 388 | E>D | No | 1000Genomes | |
| rs1967961978 | 389 | A>T | No | Ensembl | |
| rs1967962005 | 389 | A>V | No | TOPMed | |
| rs1469262399 | 390 | P>T | No | gnomAD | |
| rs1438368553 | 392 | D>Y | No |
TOPMed gnomAD |
|
| TCGA novel | 393 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1176744577 | 396 | R>C | No |
TOPMed gnomAD |
|
| rs1568353731 | 396 | R>H | No | Ensembl | |
| rs61737519 | 397 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs770598631 | 397 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs770598631 | 397 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1289064168 | 398 | L>P | No | gnomAD | |
| rs1334983274 | 400 | A>T | No | gnomAD | |
| rs1409369546 | 400 | A>V | No | gnomAD | |
| rs1347458158 | 402 | A>V | No |
TOPMed gnomAD |
|
| rs773939183 | 403 | G>S | No |
ExAC gnomAD |
|
| rs2145707610 | 404 | M>I | No | 1000Genomes | |
| rs759055479 | 404 | M>K | No |
ExAC TOPMed gnomAD |
|
| rs759055479 | 404 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1967962708 | 404 | M>V | No | gnomAD | |
| rs2145707614 | 405 | V>A | No | Ensembl | |
| rs371090751 | 406 | A>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs371090751 | 406 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1271268479 | 406 | A>V | No | gnomAD | |
| rs1199616313 | 408 | G>D | No | gnomAD | |
| rs1197594454 | 409 | P>L | No |
TOPMed gnomAD |
|
| rs1479167346 | 409 | P>S | No |
TOPMed gnomAD |
|
| rs1463428102 | 411 | G>D | No | gnomAD | |
| rs1967963122 | 411 | G>S | No | TOPMed | |
| rs762360592 | 412 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1967963206 | 412 | R>W | No | Ensembl | |
| rs1967963275 | 413 | R>C | No | TOPMed | |
| rs1425129328 | 413 | R>H | No |
TOPMed gnomAD |
|
| rs1425129328 | 413 | R>L | No |
TOPMed gnomAD |
|
| rs1425129328 | 413 | R>P | No |
TOPMed gnomAD |
|
| rs1038019512 | 414 | L>P | No | gnomAD | |
| rs1038019512 | 414 | L>Q | No | gnomAD | |
| rs896848121 | 415 | S>A | No |
TOPMed gnomAD |
|
| rs1378858516 | 415 | S>F | No | TOPMed | |
| rs896848121 | 415 | S>P | No |
TOPMed gnomAD |
|
| rs1967963597 | 417 | L>V | No |
TOPMed gnomAD |
|
| rs1196913534 | 419 | R>G | No | TOPMed | |
| rs758641980 | 419 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs758641980 | 419 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs774280689 | 420 | K>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1341876180 | 420 | K>N | No | gnomAD | |
| rs774280689 | 420 | K>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1268943880 | 420 | K>R | No | gnomAD | |
| rs1218472466 | 424 | V>L | No |
TOPMed gnomAD |
|
| TCGA novel | 426 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1038828583 | 427 | A>E | No |
TOPMed gnomAD |
|
| rs1038828583 | 427 | A>V | No |
TOPMed gnomAD |
|
| rs2145707755 | 428 | S>C | No | Ensembl | |
| rs1967963980 | 429 | T>A | No | TOPMed | |
| rs1568353832 | 429 | T>I | No |
TOPMed gnomAD |
|
| rs1967963980 | 429 | T>P | No | TOPMed | |
| rs1568353832 | 429 | T>S | No |
TOPMed gnomAD |
|
| rs1208905171 | 431 | A>S | No |
TOPMed gnomAD |
|
| rs1208905171 | 431 | A>T | No |
TOPMed gnomAD |
|
| rs755173675 | 431 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1261318181 | 433 | C>G | No | Ensembl | |
| rs1190921524 | 434 | S>* | No | gnomAD | |
| rs1190921524 | 434 | S>L | No | gnomAD | |
| rs1967964178 | 434 | S>P | No |
TOPMed gnomAD |
|
| rs1444988876 | 435 | C>* | No |
TOPMed gnomAD |
|
| rs1967964281 | 435 | C>R | No | TOPMed | |
| rs1967964281 | 435 | C>S | No | TOPMed | |
| rs1389555945 | 435 | C>Y | No | gnomAD | |
| rs781407269 | 436 | A>P | No |
ExAC gnomAD |
|
| rs781407269 | 436 | A>T | No |
ExAC gnomAD |
|
| rs1335533971 | 438 | V>F | No | TOPMed | |
| rs748082929 | 439 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs748082929 | 439 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs971251659 | 439 | P>S | No | gnomAD | |
| COSM4844103 | 440 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1365154850 | 442 | A>V | No |
TOPMed gnomAD |
|
| rs1317550646 | 443 | A>D | No |
TOPMed gnomAD |
|
| rs1317550646 | 443 | A>V | No |
TOPMed gnomAD |
|
| rs756045831 | 444 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1243542501 | 444 | P>S | No | gnomAD | |
| rs1243542501 | 444 | P>T | No | gnomAD | |
| TCGA novel | 445 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1599460011 | 445 | E>Q | No | TOPMed | |
| rs1599460012 | 446 | C>G | No | Ensembl | |
| rs777527950 | 449 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs1383077592 | 451 | P>Q | No | gnomAD | |
| TCGA novel | 453 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767255195 | 454 | D>N | No |
ExAC gnomAD |
|
| rs767255195 | 454 | D>Y | No |
ExAC gnomAD |
|
| rs1967965220 | 455 | P>L | No | Ensembl | |
| rs745509619 | 456 | G>R | No | ExAC | |
| rs745509619 | 456 | G>S | No | ExAC | |
| rs1157096928 | 457 | L>V | No | gnomAD | |
| rs1453928200 | 458 | P>L | No | gnomAD | |
| rs1453928200 | 458 | P>Q | No | gnomAD | |
| rs760066352 | 459 | E>G | No |
ExAC gnomAD |
|
| rs898722823 | 460 | P>L | No | TOPMed | |
| rs1304947735 | 461 | E>D | No |
TOPMed gnomAD |
|
| rs1307230040 | 461 | E>K | No | gnomAD | |
| rs2145707953 | 462 | A>S | No | Ensembl | |
| rs1318409072 | 464 | P>L | No |
TOPMed gnomAD |
|
| rs1967965717 | 465 | P>A | No | Ensembl | |
| rs772901494 | 465 | P>H | No |
ExAC gnomAD |
|
| rs772901494 | 465 | P>L | No |
ExAC gnomAD |
|
| rs1266898005 | 466 | A>E | No | gnomAD | |
| rs1967965822 | 466 | A>P | No | Ensembl | |
| rs1266898005 | 466 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs763379973 | 467 | G>A | No |
ExAC gnomAD |
|
| rs763379973 | 467 | G>D | No |
ExAC gnomAD |
|
| rs1568353905 | 467 | G>R | No | Ensembl | |
| rs373057256 | 469 | E>* | No |
ESP TOPMed gnomAD |
|
| rs1568353909 | 469 | E>D | No | Ensembl | |
| rs373057256 | 469 | E>K | No |
ESP TOPMed gnomAD |
|
| rs377565611 | 470 | P>Q | No |
1000Genomes ESP ExAC gnomAD |
|
| rs1251486466 | 471 | L>F | No |
TOPMed gnomAD |
|
| rs1251486466 | 471 | L>I | No |
TOPMed gnomAD |
|
| rs915230142 | 471 | L>P | No |
TOPMed gnomAD |
|
| rs1967966537 | 472 | T>I | No | Ensembl | |
| rs1967966609 | 473 | L>F | No | Ensembl | |
| rs1470379023 | 474 | I>V | No |
TOPMed gnomAD |
|
| rs978313533 | 475 | P>L | No | Ensembl | |
| rs978313533 | 475 | P>R | No | Ensembl | |
| rs752849579 | 475 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1357969573 | 476 | G>A | No |
TOPMed gnomAD |
|
| rs1357969573 | 476 | G>E | No |
TOPMed gnomAD |
|
| rs1299056002 | 477 | P>A | No | gnomAD | |
| TCGA novel | 477 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756094205 | 478 | V>D | No |
ExAC gnomAD |
|
| rs1389258446 | 478 | V>I | No | gnomAD | |
| rs777773970 | 479 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs777773970 | 479 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1599460132 | 480 | P>A | No | TOPMed | |
| rs1261139807 | 481 | F>L | No |
TOPMed gnomAD |
|
| rs1967967175 | 481 | F>S | No |
TOPMed gnomAD |
|
| rs150419376 | 482 | S>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1221722876 | 482 | S>N | No |
TOPMed gnomAD |
|
| rs1252809400 | 484 | V>L | No | gnomAD | |
| rs1967967507 | 486 | M>T | No | TOPMed | |
| rs1431217009 | 486 | M>V | No |
TOPMed gnomAD |
|
| rs1599460154 | 488 | G>R | No | Ensembl | |
| rs1180272942 | 489 | P>A | No |
TOPMed gnomAD |
|
| rs1180272942 | 489 | P>S | No |
TOPMed gnomAD |
|
| rs1251448428 | 490 | R>G | No | gnomAD | |
| rs1967967696 | 490 | R>Q | No |
TOPMed gnomAD |
|
| rs373736789 | 491 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs373736789 | 491 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1201749389 | 492 | P>L | No | gnomAD | |
| COSM3528830 | 492 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1418585189 | 492 | P>T | No |
TOPMed gnomAD |
|
| rs149797899 | 493 | A>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs745560948 | 493 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs745560948 | 493 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs941070409 | 494 | P>T | No |
TOPMed gnomAD |
|
| rs1241145188 | 495 | P>S | No | TOPMed | |
| rs1458325631 | 496 | W>S | No |
TOPMed gnomAD |
|
| rs746543024 | 499 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs2145708149 | 500 | P>L | No | Ensembl | |
| rs1437432532 | 501 | I>T | No |
TOPMed gnomAD |
|
| rs1967968225 | 501 | I>V | No | Ensembl | |
| rs1967968340 | 503 | E>V | No | Ensembl | |
| rs1406222823 | 504 | E>K | No |
TOPMed gnomAD |
|
| rs1967968392 | 505 | E>K | No | Ensembl | |
| rs1967968434 | 506 | E>K | No | TOPMed | |
| rs1357948577 | 507 | N>S | No | Ensembl | |
| rs1366326169 | 508 | L>V | No | gnomAD | |
| rs200789642 | 509 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1967968603 | 509 | A>V | No | Ensembl | |
| rs763538846 | 510 | A>R | No |
ExAC TOPMed gnomAD |
No associated diseases with Q8NFU1
5 regional properties for Q8NFU1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | AAA+ ATPase domain | 241 - 383 | IPR003593 |
| domain | ATPase, AAA-type, core | 245 - 381 | IPR003959 |
| conserved_site | ATPase, AAA-type, conserved site | 353 - 372 | IPR003960 |
| domain | Spastin/Vps4, C-terminal | 451 - 489 | IPR015415 |
| domain | AAA ATPase, AAA+ lid domain | 403 - 447 | IPR041569 |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| basolateral plasma membrane | The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis. |
| chloride channel complex | An ion channel complex through which chloride ions pass. |
| cilium | A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface and of some cytoplasmic parts. Each cilium is largely bounded by an extrusion of the cytoplasmic (plasma) membrane, and contains a regular longitudinal array of microtubules, anchored to a basal body. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| chloride channel activity | Enables the facilitated diffusion of a chloride (by an energy-independent process) involving passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism. |
| intracellularly ligand-gated monoatomic ion channel activity | Enables the transmembrane transfer of an ion by a channel that opens when a specific intracellular ligand has been bound by the channel complex or one of its constituent parts. |
| ligand-gated monoatomic anion channel activity | Enables the transmembrane transfer of an inorganic anion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts. |
| ligand-gated monoatomic cation channel activity | Enables the transmembrane transfer of an inorganic cation by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| membrane depolarization | The process in which membrane potential decreases with respect to its steady-state potential, usually from negative potential to a more positive potential. For example, the initial depolarization during the rising phase of an action potential is in the direction from the negative steady-state resting potential towards the positive membrane potential that will be the peak of the action potential. |
| sensory perception of smell | The series of events required for an organism to receive an olfactory stimulus, convert it to a molecular signal, and recognize and characterize the signal. Olfaction involves the detection of chemical composition of an organism's ambient medium by chemoreceptors. This is a neurological process. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8NFU0 | BEST4 | Bestrophin-4 | Homo sapiens (Human) | SS |
| Q8N1M1 | BEST3 | Bestrophin-3 | Homo sapiens (Human) | SS |
| O76090 | BEST1 | Bestrophin-1 | Homo sapiens (Human) | SS |
| Q6H1V1 | Best3 | Bestrophin-3 | Mus musculus (Mouse) | EV |
| O88870 | Best1 | Bestrophin-1 | Mus musculus (Mouse) | SS |
| Q8BGM5 | Best2 | Bestrophin-2 | Mus musculus (Mouse) | SS |
| O18303 | best-25 | Bestrophin homolog 25 | Caenorhabditis elegans | PR |
| O45435 | best-13 | Bestrophin homolog 13 | Caenorhabditis elegans | PR |
| P34319 | best-5 | Bestrophin homolog 5 | Caenorhabditis elegans | PR |
| P34672 | best-24 | Bestrophin homolog 24 | Caenorhabditis elegans | PR |
| Q19978 | best-14 | Bestrophin homolog 14 | Caenorhabditis elegans | PR |
| Q23369 | best-22 | Bestrophin homolog 22 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTVTYTARVA | NARFGGFSQL | LLLWRGSIYK | LLWRELLCFL | GFYMALSAAY | RFVLTEGQKR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YFEKLVIYCD | QYASLIPVSF | VLGFYVTLVV | NRWWSQYLCM | PLPDALMCVV | AGTVHGRDDR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GRLYRRTLMR | YAGLSAVLIL | RSVSTAVFKR | FPTIDHVVEA | GFMTREERKK | FENLNSSYNK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YWVPCVWFSN | LAAQARREGR | IRDNSALKLL | LEELNVFRGK | CGMLFHYDWI | SVPLVYTQVV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TIALYSYFLA | CLIGRQFLDP | AQGYKDHDLD | LCVPIFTLLQ | FFFYAGWLKV | AEQLINPFGE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DDDDFETNFL | IDRNFQVSML | AVDEMYDDLA | VLEKDLYWDA | AEARAPYTAA | TVFQLRQPSF |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QGSTFDITLA | KEDMQFQRLD | GLDGPMGEAP | GDFLQRLLPA | GAGMVAGGPL | GRRLSFLLRK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| NSCVSEASTG | ASCSCAVVPE | GAAPECSCGD | PLLDPGLPEP | EAPPPAGPEP | LTLIPGPVEP |
| 490 | 500 | ||||
| FSIVTMPGPR | GPAPPWLPSP | IGEEEENLA |