Q8NCB2
Gene name |
CAMKV |
Protein name |
CaM kinase-like vesicle-associated protein |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:79012 |
EC number |
|
Protein Class |
|
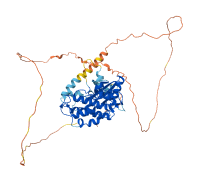
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
24-286 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
165-185 (Activation loop from InterPro)
Target domain |
24-286 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Goldberg J et al. (1996) "Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I", Cell, 84, 875-87
- Clapperton JA et al. (2002) "Structure of the complex of calmodulin with the target sequence of calmodulin-dependent protein kinase I: studies of the kinase activation mechanism", Biochemistry, 41, 14669-79
- Yuan T et al. (2004) "Spectroscopic characterization of the calmodulin-binding and autoinhibitory domains of calcium/calmodulin-dependent protein kinase I", Archives of biochemistry and biophysics, 421, 192-206
- Zha M et al. (2012) "Crystal structures of human CaMKIα reveal insights into the regulation mechanism of CaMKI", PloS one, 7, e44828
- Shohat G et al. (2002) "The DAP-kinase family of proteins: study of a novel group of calcium-regulated death-promoting kinases", Biochimica et biophysica acta, 1600, 45-50
- Cohen O et al. (1997) "DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity", The EMBO journal, 16, 998-1008
- Shohat G et al. (2001) "The pro-apoptotic function of death-associated protein kinase is controlled by a unique inhibitory autophosphorylation-based mechanism", The Journal of biological chemistry, 276, 47460-7
- Temmerman K et al. (2014) "A PEF/Y substrate recognition and signature motif plays a critical role in DAPK-related kinase activity", Chemistry & biology, 21, 264-73
Autoinhibited structure

Activated structure

1 structures for Q8NCB2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8NCB2-F1 | Predicted | AlphaFoldDB |
376 variants for Q8NCB2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs2082029117 | 2 | P>S | No | Ensembl | |
| TCGA novel | 8 | L>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4118636 COSM4118637 |
8 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs147683930 | 8 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2108330269 | 10 | D>N | No | Ensembl | |
| rs1302561608 | 11 | K>E | No |
TOPMed gnomAD |
|
|
rs771453537 COSM1046326 COSM1046325 |
12 | K>N | endometrium Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated ExAC gnomAD NCI-TCGA Cosmic |
| rs1176714251 | 12 | K>R | No |
TOPMed gnomAD |
|
| rs2108330260 | 13 | N>Y | No | Ensembl | |
| rs1185840055 | 17 | P>T | No | gnomAD | |
| rs1575407453 | 20 | V>G | No | Ensembl | |
|
COSM446730 COSM1485321 |
20 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3595442 COSM3595443 |
25 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1312975289 | 30 | I>M | No |
TOPMed gnomAD |
|
| rs2082028420 | 31 | K>T | No | TOPMed | |
| rs369471161 | 32 | T>I | No |
ESP TOPMed |
|
| TCGA novel | 36 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
VAR_041337 rs1407808379 |
40 | R>W | a colorectal adenocarcinoma sample; somatic mutation [UniProt] | No |
UniProt dbSNP gnomAD |
| rs1426803266 | 41 | A>V | No | gnomAD | |
| rs1241766696 | 43 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| TCGA novel | 44 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747162209 | 45 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2082026770 | 47 | G>D | No | TOPMed | |
| rs1443985656 | 48 | K>R | No |
TOPMed gnomAD |
|
| rs765666578 | 49 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs2082026707 | 49 | L>V | No | TOPMed | |
| rs757504862 | 51 | T>N | No |
ExAC gnomAD |
|
| TCGA novel | 52 | C>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1250012626 | 54 | K>M | No | gnomAD | |
| rs774049687 | 58 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs200718388 | 58 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
VAR_041338 COSM20715 rs2082026503 COSM4118635 |
60 | G>S | ovary Variant assessed as Somatic; MODERATE impact. an ovarian serous carcinoma sample; somatic mutation [Cosmic, NCI-TCGA, UniProt] | No |
NCI-TCGA Cosmic cosmic curated TOPMed UniProt |
| rs1360882177 | 61 | R>C | No |
TOPMed gnomAD |
|
| rs772689036 | 61 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1575407229 | 63 | V>G | No | Ensembl | |
| rs1238361476 | 64 | R>Q | No | gnomAD | |
| rs1305797638 | 68 | K>E | No | gnomAD | |
|
rs865944634 COSM5603061 COSM5603060 |
70 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs769648151 | 71 | I>M | No |
ExAC gnomAD |
|
| rs1029418614 | 72 | G>A | No | Ensembl | |
| rs1257316947 | 72 | G>S | No | Ensembl | |
|
COSM3373234 COSM3373233 |
75 | K>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM1046323 COSM1046324 |
75 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1374844644 | 75 | K>R | No | gnomAD | |
| TCGA novel | 76 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1575407017 | 77 | V>G | No | Ensembl | |
| rs1446975655 | 79 | H>Q | No | gnomAD | |
| rs1262864897 | 82 | I>V | No | gnomAD | |
| rs2108329827 | 84 | Q>R | No | Ensembl | |
| rs144366691 | 85 | L>P | No | ESP | |
| rs1482489474 | 86 | V>M | No | gnomAD | |
| rs1301880158 | 87 | D>N | No | gnomAD | |
|
COSM74002 COSM74001 |
92 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1344113248 | 92 | R>L | No |
TOPMed gnomAD |
|
| rs1559456830 | 93 | K>E | No | Ensembl | |
| rs2082024156 | 93 | K>R | No | TOPMed | |
| TCGA novel | 95 | Y>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4811679 COSM419940 |
97 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1284693414 | 97 | I>S | No | gnomAD | |
| TCGA novel | 99 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746000364 | 99 | L>Q | No |
ExAC gnomAD |
|
| rs866173336 | 101 | L>M | No | gnomAD | |
| rs2082021644 | 102 | A>V | No | TOPMed | |
| rs760406345 | 103 | T>M | No |
ExAC gnomAD |
|
| rs1575406791 | 107 | V>G | No | Ensembl | |
| rs2082021440 | 107 | V>L | No | TOPMed | |
| rs2082021333 | 109 | D>E | No | TOPMed | |
| rs2082021295 | 110 | W>C | No | Ensembl | |
|
COSM3916295 COSM3916294 |
113 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs866912622 | 115 | G>S | No | Ensembl | |
| rs2082021183 | 115 | G>V | No | Ensembl | |
| COSM1424054 | 116 | Y>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1288269620 | 117 | Y>H | No | TOPMed | |
|
COSM1485320 COSM1485319 |
118 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4118631 COSM4118632 |
119 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1384999199 | 120 | R>Q | No | gnomAD | |
| TCGA novel | 122 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2108329596 | 123 | S>C | No | Ensembl | |
|
COSM1046322 COSM1046321 |
125 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1446833091 | 126 | V>I | No | gnomAD | |
| rs2082020750 | 127 | R>Q | No | TOPMed | |
| rs774334647 | 127 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs770998448 | 129 | V>I | No |
ExAC gnomAD |
|
| rs1273162438 | 130 | L>P | No | gnomAD | |
| rs1176759665 | 133 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs770285366 | 134 | A>D | No |
ExAC gnomAD |
|
| rs1290746713 | 134 | A>S | No | gnomAD | |
| rs770285366 | 134 | A>V | No |
ExAC gnomAD |
|
| TCGA novel | 136 | L>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4118630 COSM4118629 |
141 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 142 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 144 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2082020370 | 147 | K>Q | No | Ensembl | |
| COSM274371 | 149 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1261033358 | 152 | V>L | No |
TOPMed gnomAD |
|
| rs770201843 | 154 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs1198915352 | 155 | N>S | No | TOPMed | |
| rs1251987183 | 156 | R>Q | No | TOPMed | |
| rs777179632 | 156 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs769025060 | 157 | L>M | No | ExAC | |
| rs2082018662 | 158 | K>R | No | Ensembl | |
| rs2082018647 | 160 | S>L | No | Ensembl | |
| rs2082018417 | 175 | G>D | No | TOPMed | |
| rs777241750 | 177 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs2082018353 | 181 | C>* | No | Ensembl | |
| rs2108329340 | 184 | P>H | No | Ensembl | |
| rs1300168935 | 185 | E>D | No | TOPMed | |
| rs1297165732 | 189 | P>Q | No | gnomAD | |
| rs190982523 | 191 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA |
|
COSM1046319 COSM1046318 rs2082015579 |
193 | G>D | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
|
COSM3916292 COSM3916293 |
193 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1410351118 | 194 | R>Q | No | gnomAD | |
|
COSM1642299 rs1361038100 COSM1642298 |
194 | R>W | stomach [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| TCGA novel | 195 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760054653 | 196 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs1220869354 COSM1424052 COSM1424051 |
196 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs1161602649 | 197 | Y>C | No | gnomAD | |
| rs1575406359 | 197 | Y>D | No | Ensembl | |
| rs774782174 | 199 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs769296484 | 199 | R>H | No |
ExAC gnomAD |
|
|
COSM3595434 COSM3595435 |
200 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3595432 COSM3595433 |
205 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs966029827 | 206 | I>V | No |
TOPMed gnomAD |
|
| TCGA novel | 207 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747722227 | 210 | M>T | No |
ExAC gnomAD |
|
|
COSM3595430 COSM3595431 |
212 | I>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM1046314 rs200019805 COSM1046315 |
213 | L>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
|
COSM1424050 COSM1424049 |
213 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2108328998 | 216 | G>S | No | Ensembl | |
| rs1317972592 | 227 | D>E | No |
TOPMed gnomAD |
|
| rs1199285295 | 227 | D>N | No |
TOPMed gnomAD |
|
| rs1199285295 | 227 | D>Y | No |
TOPMed gnomAD |
|
| TCGA novel | 230 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2082013700 | 231 | N>S | No | Ensembl | |
|
COSM3916289 COSM3916288 |
233 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3373232 COSM3373231 |
234 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1269893469 | 235 | N>T | No | gnomAD | |
|
COSM3595429 COSM3595428 |
236 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763349594 | 238 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs2082013603 TCGA novel |
238 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1485750027 | 242 | A>V | No |
TOPMed gnomAD |
|
| rs776246810 | 248 | D>E | No |
ExAC TOPMed gnomAD |
|
|
COSM6165094 COSM6165095 |
250 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1245270711 | 251 | Y>H | No | TOPMed | |
| TCGA novel | 252 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768011837 | 254 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs768011837 | 254 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1442840395 | 254 | D>Y | No | Ensembl | |
| rs143595972 | 260 | K>R | No |
ESP TOPMed gnomAD |
|
| TCGA novel | 263 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1418449261 | 264 | T>I | No | TOPMed | |
| rs2082011813 | 265 | R>K | No | TOPMed | |
|
COSM3595427 COSM3595426 |
268 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2082011735 | 269 | V>M | No |
TOPMed gnomAD |
|
| VAR_041339 | 274 | R>W | a colorectal adenocarcinoma sample; somatic mutation [UniProt] | No | UniProt |
| rs1240837292 | 278 | E>K | No | gnomAD | |
|
VAR_041340 rs56071455 |
279 | E>D | No |
UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs2082011597 | 281 | I>V | No |
TOPMed gnomAD |
|
| rs944287748 | 284 | E>D | No |
TOPMed gnomAD |
|
| rs2082011524 | 284 | E>G | No | Ensembl | |
|
COSM6098027 COSM6098028 |
292 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2082009694 | 293 | D>E | No | Ensembl | |
| TCGA novel | 293 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2082009725 | 293 | D>V | No | TOPMed | |
| rs759280881 | 297 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs143217264 | 299 | G>A | No |
ESP ExAC gnomAD |
|
| rs143217264 | 299 | G>D | No |
ESP ExAC gnomAD |
|
| rs762722820 | 303 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
|
TCGA novel rs2082009609 |
304 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
|
COSM1046311 COSM1046310 |
306 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2082009546 | 309 | A>V | No | Ensembl | |
| COSM266180 | 310 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2082009528 | 311 | A>T | No | Ensembl | |
| rs900943884 | 314 | K>R | No | Ensembl | |
| TCGA novel | 316 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1424296949 | 318 | R>* | No | gnomAD | |
| rs1191733979 | 318 | R>Q | No | gnomAD | |
| rs2082006677 | 325 | R>Q | No | TOPMed | |
| rs779576572 | 325 | R>W | No |
ExAC gnomAD |
|
| rs2082006639 | 327 | R>Q | No | gnomAD | |
| rs573710169 | 327 | R>W | No |
1000Genomes gnomAD |
|
| rs772919486 | 328 | A>T | No |
ExAC gnomAD |
|
| rs917359050 | 330 | E>K | No |
TOPMed gnomAD |
|
| rs1267260615 | 331 | Q>R | No | TOPMed | |
| rs961401656 | 332 | S>C | No | gnomAD | |
| rs961401656 | 332 | S>Y | No | gnomAD | |
| rs1017181631 | 333 | S>G | No |
TOPMed gnomAD |
|
| rs1360279426 | 333 | S>N | No | gnomAD | |
| rs1017181631 | 333 | S>R | No |
TOPMed gnomAD |
|
| rs376450781 | 334 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 335 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs984737561 | 335 | A>V | No | Ensembl | |
| rs768728034 | 336 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1358119570 | 337 | A>G | No |
TOPMed gnomAD |
|
| rs1467739973 | 337 | A>S | No | gnomAD | |
| rs1559455873 | 338 | Q>E | No | Ensembl | |
| rs1416168874 | 338 | Q>L | No |
TOPMed gnomAD |
|
| rs1416168874 | 338 | Q>R | No |
TOPMed gnomAD |
|
| rs780173738 | 339 | S>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs746074470 | 343 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1471051101 | 345 | T>A | No | TOPMed | |
| rs1250087757 | 346 | A>T | No |
TOPMed gnomAD |
|
| rs779164652 | 346 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1238167908 | 347 | T>A | No | gnomAD | |
| rs1212860694 | 347 | T>I | No | gnomAD | |
| rs1238167908 | 347 | T>P | No | gnomAD | |
| TCGA novel | 348 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2082005360 | 348 | P>T | No | TOPMed | |
| rs749812937 | 349 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1229510660 | 351 | A>E | No |
TOPMed gnomAD |
|
| rs1229510660 | 351 | A>V | No |
TOPMed gnomAD |
|
| rs1344797857 | 353 | G>R | No | gnomAD | |
| rs1337387637 | 354 | A>P | No |
TOPMed gnomAD |
|
| rs1337387637 | 354 | A>T | No |
TOPMed gnomAD |
|
| rs756675433 | 355 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1377801438 | 355 | T>I | No | gnomAD | |
| rs1302979931 | 357 | A>P | No | gnomAD | |
|
rs1368553470 COSM339151 |
358 | A>D | lung [Cosmic] | No |
cosmic curated gnomAD |
| rs1449602905 | 358 | A>P | No |
TOPMed gnomAD |
|
| rs1307371534 | 359 | A>E | No |
TOPMed gnomAD |
|
| rs1307371534 | 359 | A>V | No |
TOPMed gnomAD |
|
| rs1575405483 | 360 | S>I | No | Ensembl | |
| rs2082005024 | 360 | S>R | No |
TOPMed gnomAD |
|
| rs765879384 | 361 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs2082004964 | 361 | G>V | No |
TOPMed gnomAD |
|
| rs758016459 | 362 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1482637734 | 362 | A>V | No | TOPMed | |
| rs149916894 | 365 | A>V | No |
1000Genomes ESP TOPMed |
|
| rs2108328153 | 368 | G>V | No | Ensembl | |
| rs1414614976 | 370 | A>T | No | gnomAD | |
|
rs377281206 COSM4459166 COSM4459167 |
372 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM5183747 rs199584164 |
372 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs199584164 | 372 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2082004642 | 373 | A>T | No | TOPMed | |
| rs1206531994 | 375 | K>E | No | gnomAD | |
| rs1020690637 | 375 | K>N | No | gnomAD | |
| rs2082004578 | 378 | N>K | No | Ensembl | |
| rs1239415454 | 379 | V>A | No |
TOPMed gnomAD |
|
| rs1239415454 | 379 | V>E | No |
TOPMed gnomAD |
|
| rs1290123705 | 379 | V>M | No | gnomAD | |
| rs764079591 | 380 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs764079591 | 380 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs868127460 | 381 | P>S | No | Ensembl | |
| rs772138978 | 382 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs150720881 | 382 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs772138978 | 382 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1053448441 | 384 | R>C | No |
TOPMed gnomAD |
|
| rs746089971 | 384 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs746089971 | 384 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM6165097 COSM6165096 COSM1538195 rs2082004293 COSM1538196 |
385 | S>N | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic TOPMed |
| rs774562098 | 387 | T>P | No |
ExAC gnomAD |
|
| rs1249255895 | 387 | T>S | No | gnomAD | |
| rs142041712 | 388 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs749727173 | 390 | T>R | No |
ExAC gnomAD |
|
| rs934957635 | 391 | D>E | No |
TOPMed gnomAD |
|
|
COSM3595421 COSM3595420 |
391 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs866327852 | 394 | A>V | No | Ensembl | |
| rs778259877 | 395 | T>P | No |
ExAC gnomAD |
|
| rs748648023 | 396 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs2108328059 | 396 | P>T | No | Ensembl | |
| rs1559455721 | 399 | D>G | No |
TOPMed gnomAD |
|
| rs2108328044 | 399 | D>H | No | Ensembl | |
| rs757869703 | 400 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs556114266 | 401 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2082003752 | 402 | V>A | No | Ensembl | |
| rs1265610524 | 403 | T>N | No | gnomAD | |
| rs2108328031 | 403 | T>P | No | Ensembl | |
| rs2082003709 | 404 | P>S | No |
TOPMed gnomAD |
|
| rs1212043537 | 405 | A>V | No | gnomAD | |
| rs1326421924 | 407 | D>E | No | gnomAD | |
|
COSM4118626 COSM4118625 rs756893325 |
407 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1228920749 | 410 | I>V | No |
TOPMed gnomAD |
|
| rs1352197417 | 411 | T>A | No | gnomAD | |
| rs1195660596 | 411 | T>I | No | TOPMed | |
| rs1352197417 | 411 | T>P | No | gnomAD | |
| rs763848470 | 412 | P>L | No |
ExAC gnomAD |
|
| rs1233862924 | 412 | P>S | No | Ensembl | |
| rs760641215 | 413 | A>T | No |
ExAC gnomAD |
|
| rs2082003280 | 415 | D>E | No | TOPMed | |
|
rs2082003250 COSM141045 |
416 | G>E | upper_aerodigestive_tract [Cosmic] | No |
cosmic curated Ensembl |
| rs200719095 | 416 | G>R | No |
1000Genomes ExAC |
|
| rs201137978 | 417 | S>N | No |
1000Genomes TOPMed gnomAD |
|
| rs1387977257 | 419 | T>N | No | gnomAD | |
| rs774277628 | 419 | T>P | No |
ExAC gnomAD |
|
| rs1311611744 | 420 | P>L | No | gnomAD | |
| rs143724249 | 423 | D>G | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1046308 COSM1046309 |
424 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2082003024 | 425 | S>G | No | TOPMed | |
| rs773741414 | 426 | A>P | No |
ExAC gnomAD |
|
| rs773741414 | 426 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2082002955 | 426 | A>V | No | gnomAD | |
| rs770232583 | 427 | T>P | No |
ExAC gnomAD |
|
| rs1002498837 | 428 | P>L | No |
TOPMed gnomAD |
|
| rs781676831 | 428 | P>S | No |
ExAC gnomAD |
|
|
COSM3696162 COSM3696163 |
429 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3916287 COSM3916286 rs769039667 |
429 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs778376639 | 430 | T>N | No |
ExAC gnomAD |
|
| rs1303210385 | 433 | R>K | No |
TOPMed gnomAD |
|
| rs2082002702 | 433 | R>S | No | Ensembl | |
| rs905124046 | 434 | A>D | No | Ensembl | |
| rs756948275 | 434 | A>P | No |
ExAC gnomAD |
|
| rs905124046 | 434 | A>V | No | Ensembl | |
| rs753451563 | 435 | T>P | No |
ExAC gnomAD |
|
| rs2082002533 | 437 | A>V | No |
TOPMed gnomAD |
|
| rs201726498 | 438 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2082002510 | 438 | T>S | No | TOPMed | |
| rs1173582479 | 439 | E>K | No |
TOPMed gnomAD |
|
| rs144523890 | 440 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs2082002394 | 443 | V>A | No | TOPMed | |
| rs767544686 | 443 | V>M | No |
ExAC gnomAD |
|
| rs917329705 | 444 | P>L | No | Ensembl | |
| rs1449247412 | 445 | T>N | No | gnomAD | |
| rs1575405152 | 445 | T>P | No | Ensembl | |
| rs1449247412 | 445 | T>S | No | gnomAD | |
| rs1188833621 | 446 | T>N | No | gnomAD | |
| rs549068466 | 446 | T>P | No | 1000Genomes | |
| rs2082002127 | 449 | S>G | No | TOPMed | |
| rs1199790578 | 450 | A>T | No | gnomAD | |
| rs2108327846 | 451 | M>T | No | Ensembl | |
| rs2082002086 | 452 | L>M | No | TOPMed | |
| rs2082002069 | 453 | A>T | No | TOPMed | |
| rs1265199404 | 454 | T>A | No |
TOPMed gnomAD |
|
| rs1026894871 | 455 | K>R | No |
TOPMed gnomAD |
|
| rs1259989462 | 456 | A>S | No | Ensembl | |
| rs1341226202 | 459 | T>N | No | gnomAD | |
| rs1575405106 | 459 | T>P | No | TOPMed | |
| TCGA novel | 460 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM6098029 COSM6098030 |
460 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs199527105 | 462 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199527105 | 462 | P>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs867560885 | 463 | A>T | No |
TOPMed gnomAD |
|
| rs1446319423 | 464 | M>T | No |
TOPMed gnomAD |
|
| rs1338769404 | 464 | M>V | No | gnomAD | |
| rs762305255 | 467 | P>L | No |
ExAC TOPMed gnomAD |
|
|
COSM3408753 COSM3408754 |
467 | P>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1454180733 | 468 | D>G | No |
TOPMed gnomAD |
|
|
COSM3595419 COSM3595418 |
468 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1404898456 | 470 | T>A | No |
TOPMed gnomAD |
|
| rs2082001661 | 470 | T>I | No |
TOPMed gnomAD |
|
| rs1306753877 | 471 | A>D | No |
TOPMed gnomAD |
|
| rs1306753877 | 471 | A>G | No |
TOPMed gnomAD |
|
|
VAR_041341 rs56307047 |
472 | P>L | No |
UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1429463958 | 472 | P>S | No |
TOPMed gnomAD |
|
| rs1429463958 | 472 | P>T | No |
TOPMed gnomAD |
|
| TCGA novel | 474 | G>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2082001541 | 474 | G>S | No | TOPMed | |
| rs770382119 | 475 | A>D | No |
ExAC gnomAD |
|
| rs778427920 | 475 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs748940995 | 477 | G>A | No |
ExAC gnomAD |
|
| rs748940995 | 477 | G>D | No |
ExAC gnomAD |
|
|
COSM1046306 COSM1046307 |
479 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs778514209 | 479 | A>V | No |
TOPMed gnomAD |
|
| rs1208427176 | 480 | P>S | No |
TOPMed gnomAD |
|
| rs1482587570 | 481 | P>H | No | gnomAD | |
| rs1482587570 | 481 | P>L | No | gnomAD | |
| rs1011785677 | 483 | S>G | No |
TOPMed gnomAD |
|
| rs1011785677 | 483 | S>R | No |
TOPMed gnomAD |
|
| rs893317238 | 483 | S>T | No |
TOPMed gnomAD |
|
| rs777466315 | 486 | E>K | No |
ExAC gnomAD |
|
| rs752582179 | 489 | A>P | No |
ExAC gnomAD |
|
| rs1245564647 | 490 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
VAR_027539 rs17849325 |
491 | Y>C | No |
UniProt Ensembl dbSNP |
|
| rs1286982887 | 492 | A>S | No |
TOPMed gnomAD |
|
| rs1286982887 | 492 | A>T | No |
TOPMed gnomAD |
|
| rs1409795216 | 492 | A>V | No | gnomAD | |
| rs754974030 | 494 | E>A | No |
ExAC gnomAD |
|
| rs1371556597 | 494 | E>Q | No |
TOPMed gnomAD |
|
| rs1403956518 | 496 | Q>E | No | gnomAD | |
| rs751491503 | 496 | Q>P | No |
ExAC gnomAD |
|
|
COSM4169792 COSM4169791 |
497 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1472827918 | 497 | R>S | No | gnomAD | |
| rs149379959 | 497 | R>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs757078390 | 498 | E>V | No | Ensembl | |
| rs2082000886 | 499 | E>G | No | TOPMed | |
| rs2082000869 | 500 | A>D | No | TOPMed | |
|
COSM6165099 COSM6165098 |
501 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
No associated diseases with Q8NCB2
No regional properties for Q8NCB2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q8NCB2 | |||
Functions
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasmic vesicle membrane | The lipid bilayer surrounding a cytoplasmic vesicle. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein kinase activity | Calmodulin-dependent catalysis of the reactions |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| modulation of chemical synaptic transmission | Any process that modulates the frequency or amplitude of synaptic transmission, the process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse. Amplitude, in this case, refers to the change in postsynaptic membrane potential due to a single instance of synaptic transmission. |
| regulation of modification of postsynaptic structure | Any process that modulates the frequency, rate or extent of modification of postsynaptic structure. |
106 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A4IFM7 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Bos taurus (Bovine) | SS |
| Q0V7M1 | PSKH1 | Serine/threonine-protein kinase H1 | Bos taurus (Bovine) | SS |
| Q0VD22 | STK33 | Serine/threonine-protein kinase 33 | Bos taurus (Bovine) | PR |
| O15075 | DCLK1 | Serine/threonine-protein kinase DCLK1 | Homo sapiens (Human) | EV |
| Q9UIK4 | DAPK2 | Death-associated protein kinase 2 | Homo sapiens (Human) | EV |
| O94768 | STK17B | Serine/threonine-protein kinase 17B | Homo sapiens (Human) | PR |
| Q16566 | CAMK4 | Calcium/calmodulin-dependent protein kinase type IV | Homo sapiens (Human) | SS |
| Q8N568 | DCLK2 | Serine/threonine-protein kinase DCLK2 | Homo sapiens (Human) | PR |
| Q9UEE5 | STK17A | Serine/threonine-protein kinase 17A | Homo sapiens (Human) | SS |
| O75962 | TRIO | Triple functional domain protein | Homo sapiens (Human) | EV |
| Q8WWF8 | CAPSL | Calcyphosin-like protein | Homo sapiens (Human) | PR |
| P53355 | DAPK1 | Death-associated protein kinase 1 | Homo sapiens (Human) | EV |
| Q9BYT3 | STK33 | Serine/threonine-protein kinase 33 | Homo sapiens (Human) | PR |
| O43293 | DAPK3 | Death-associated protein kinase 3 | Homo sapiens (Human) | PR |
| Q16816 | PHKG1 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform | Homo sapiens (Human) | PR |
| P15735 | PHKG2 | Phosphorylase b kinase gamma catalytic chain, liver/testis isoform | Homo sapiens (Human) | PR |
| Q13554 | CAMK2B | Calcium/calmodulin-dependent protein kinase type II subunit beta | Homo sapiens (Human) | EV |
| Q9UQM7 | CAMK2A | Calcium/calmodulin-dependent protein kinase type II subunit alpha | Homo sapiens (Human) | EV |
| Q13555 | CAMK2G | Calcium/calmodulin-dependent protein kinase type II subunit gamma | Homo sapiens (Human) | EV |
| Q13557 | CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta | Homo sapiens (Human) | EV |
| Q9H1R3 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Homo sapiens (Human) | EV |
| Q32MK0 | MYLK3 | Myosin light chain kinase 3 | Homo sapiens (Human) | SS |
| Q86YV6 | MYLK4 | Myosin light chain kinase family member 4 | Homo sapiens (Human) | SS |
| P11801 | PSKH1 | Serine/threonine-protein kinase H1 | Homo sapiens (Human) | SS |
| Q6P2M8 | PNCK | Calcium/calmodulin-dependent protein kinase type 1B | Homo sapiens (Human) | SS |
| Q96NX5 | CAMK1G | Calcium/calmodulin-dependent protein kinase type 1G | Homo sapiens (Human) | SS |
| Q8IU85 | CAMK1D | Calcium/calmodulin-dependent protein kinase type 1D | Homo sapiens (Human) | SS |
| Q14012 | CAMK1 | Calcium/calmodulin-dependent protein kinase type 1 | Homo sapiens (Human) | EV |
| Q9QYK9 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Mus musculus (Mouse) | PR |
| Q91VB2 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Mus musculus (Mouse) | SS |
| Q8BG48 | Stk17b | Serine/threonine-protein kinase 17B | Mus musculus (Mouse) | PR |
| Q8VCR8 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Mus musculus (Mouse) | SS |
| Q6PGN3 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Mus musculus (Mouse) | PR |
| Q91YS8 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Mus musculus (Mouse) | SS |
| Q3UIZ8 | Mylk3 | Myosin light chain kinase 3 | Mus musculus (Mouse) | SS |
| Q8VDF3 | Dapk2 | Death-associated protein kinase 2 | Mus musculus (Mouse) | EV |
| Q6P8Y1 | Capsl | Calcyphosin-like protein | Mus musculus (Mouse) | PR |
| Q8BW96 | Camk1d | Calcium/calmodulin-dependent protein kinase type 1D | Mus musculus (Mouse) | SS |
| P08414 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Mus musculus (Mouse) | SS |
| Q9JLM8 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Mus musculus (Mouse) | SS |
| Q91YA2 | Pskh1 | Serine/threonine-protein kinase H1 | Mus musculus (Mouse) | SS |
| Q80YE7 | Dapk1 | Death-associated protein kinase 1 | Mus musculus (Mouse) | SS |
| Q62407 | Speg | Striated muscle-specific serine/threonine-protein kinase | Mus musculus (Mouse) | SS |
| Q0KL02 | Trio | Triple functional domain protein | Mus musculus (Mouse) | SS |
| Q924X7 | Stk33 | Serine/threonine-protein kinase 33 | Mus musculus (Mouse) | PR |
| O54784 | Dapk3 | Death-associated protein kinase 3 | Mus musculus (Mouse) | PR |
| Q3UHL1 | Camkv | CaM kinase-like vesicle-associated protein | Mus musculus (Mouse) | SS |
| Q7TNJ7 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Rattus norvegicus (Rat) | SS |
| P20689 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Rattus norvegicus (Rat) | SS |
| F1M0Z1 | Trio | Triple functional domain protein | Rattus norvegicus (Rat) | SS |
| Q91XS8 | Stk17b | Serine/threonine-protein kinase 17B | Rattus norvegicus (Rat) | PR |
| O08875 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Rattus norvegicus (Rat) | PR |
| E9PT87 | Mylk3 | Myosin light chain kinase 3 | Rattus norvegicus (Rat) | SS |
| O70150 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Rattus norvegicus (Rat) | PR |
| Q63638 | Speg | Striated muscle-specific serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| O88764 | Dapk3 | Death-associated protein kinase 3 | Rattus norvegicus (Rat) | PR |
| P13234 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Rattus norvegicus (Rat) | SS |
| Q63450 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Rattus norvegicus (Rat) | EV |
| P97924 | Kalrn | Kalirin | Rattus norvegicus (Rat) | SS |
| Q5MPA9 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Rattus norvegicus (Rat) | PR |
| Q63092 | Camkv | CaM kinase-like vesicle-associated protein | Rattus norvegicus (Rat) | SS |
| Q6AVM3 | CCAMK | Calcium and calcium/calmodulin-dependent serine/threonine-protein kinase | Oryza sativa subsp japonica (Rice) | PR |
| P53682 | CPK23 | Calcium-dependent protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| P53684 | CPK7 | Calcium-dependent protein kinase 7 | Oryza sativa subsp japonica (Rice) | PR |
| Q5VQQ5 | CPK2 | Calcium-dependent protein kinase 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6I5I8 | CPK16 | Calcium-dependent protein kinase 16 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6K968 | CPK6 | Calcium-dependent protein kinase 6 | Oryza sativa subsp. japonica (Rice) | SS |
| Q75GE8 | CPK8 | Calcium-dependent protein kinase 8 | Oryza sativa subsp. japonica (Rice) | SS |
| Q84SL0 | CPK20 | Calcium-dependent protein kinase 20 | Oryza sativa subsp. japonica (Rice) | SS |
| Q8LPZ7 | CPK3 | Calcium-dependent protein kinase 3 | Oryza sativa subsp. japonica (Rice) | SS |
| O44997 | dapk-1 | Death-associated protein kinase dapk-1 | Caenorhabditis elegans | SS |
| Q95QC4 | zyg-8 | Serine/threonine-protein kinase zyg-8 | Caenorhabditis elegans | PR |
| Q9TXJ0 | cmk-1 | Calcium/calmodulin-dependent protein kinase type 1 | Caenorhabditis elegans | PR |
| P28583 | Calcium-dependent protein kinase SK5 | Glycine max (Soybean) (Glycine hispida) | PR | |
| Q9M9V8 | CPK10 | Calcium-dependent protein kinase 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FX86 | CRK8 | CDPK-related kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SSF8 | CPK30 | Calcium-dependent protein kinase 30 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIQ7 | CPK24 | Calcium-dependent protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93759 | GK-1 | Calcium-dependent protein kinase 14 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8S8S2 | LPEAT2 | Lysophospholipid acyltransferase LPEAT2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LJL9 | CRK2 | CDPK-related kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SG12 | CRK6 | CDPK-related kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCS2 | CRK5 | CDPK-related kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8W4I7 | CPK13 | Calcium-dependent protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NLQ6 | CPK32 | Calcium-dependent protein kinase 32 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38873 | CPK7 | Calcium-dependent protein kinase 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42438 | CPK8 | Calcium-dependent protein kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q06850 | CPK1 | Calcium-dependent protein kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38869 | CPK4 | Calcium-dependent protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38870 | CPK2 | Calcium-dependent protein kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38872 | CPK6 | Calcium-dependent protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39016 | CPK11 | Calcium-dependent protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E9C0 | CPK34 | Calcium-dependent protein kinase 34 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42396 | CPK12 | Calcium-dependent protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42479 | CPK3 | Calcium-dependent protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7XJR9 | CPK16 | Calcium-dependent protein kinase 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FMP5 | CPK17 | Calcium-dependent protein kinase 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJ61 | CPK25 | Calcium-dependent protein kinase 25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZSA4 | CPK27 | Calcium-dependent protein kinase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZV15 | CPK20 | Calcium-dependent protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q1PE17 | CPK18 | Calcium-dependent protein kinase 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FKW4 | CPK28 | Calcium-dependent protein kinase 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1LUA6 | trio | Triple functional domain protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A8C984 | mylk3 | Myosin light chain kinase 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q7SY49 | camkv | CaM kinase-like vesicle-associated protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPFGCVTLGD | KKNYNQPSEV | TDRYDLGQVI | KTEEFCEIFR | AKDKTTGKLH | TCKKFQKRDG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RKVRKAAKNE | IGILKMVKHP | NILQLVDVFV | TRKEYFIFLE | LATGREVFDW | ILDQGYYSER |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DTSNVVRQVL | EAVAYLHSLK | IVHRNLKLEN | LVYYNRLKNS | KIVISDFHLA | KLENGLIKEP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| CGTPEYLAPE | VVGRQRYGRP | VDCWAIGVIM | YILLSGNPPF | YEEVEEDDYE | NHDKNLFRKI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LAGDYEFDSP | YWDDISQAAK | DLVTRLMEVE | QDQRITAEEA | ISHEWISGNA | ASDKNIKDGV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| CAQIEKNFAR | AKWKKAVRVT | TLMKRLRAPE | QSSTAAAQSA | SATDTATPGA | AGGATAAAAS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GATSAPEGDA | ARAAKSDNVA | PADRSATPAT | DGSATPATDG | SVTPATDGSI | TPATDGSVTP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| ATDRSATPAT | DGRATPATEE | STVPTTQSSA | MLATKAAATP | EPAMAQPDST | APEGATGQAP |
| 490 | 500 | ||||
| PSSKGEEAAG | YAQESQREEA | S |