Q8N831
Gene name |
TSPYL6 |
Protein name |
Testis-specific Y-encoded-like protein 6 |
Names |
TSPY-like protein 6 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:388951 |
EC number |
|
Protein Class |
|
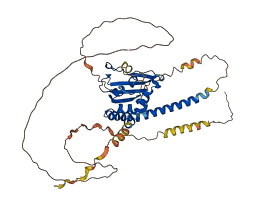
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q8N831
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8N831-F1 | Predicted | AlphaFoldDB |
50 variants for Q8N831
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000973303 rs148872913 |
1 | M>T | No |
ClinVar dbSNP |
|
|
rs61736720 RCV000953241 |
1 | M>V | No |
ClinVar dbSNP |
|
|
rs113932278 RCV000968542 |
4 | P>L | No |
ClinVar dbSNP |
|
| rs1431602063 | 11 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 39 | M>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs537805712 | 43 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs537805712 | 43 | R>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV000906665 rs61732717 |
51 | P>L | No |
ClinVar dbSNP |
|
| rs1309150915 | 57 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 59 | V>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs6743719 RCV000969431 VAR_035395 |
60 | A>V | No |
ClinVar UniProt dbSNP |
|
| TCGA novel | 72 | H>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs372025284 | 76 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 77 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746897592 | 79 | R>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776617090 | 95 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs372159200 | 95 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV000953240 rs61738381 |
96 | E>D | No |
ClinVar dbSNP |
|
| rs1243685150 | 100 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
VAR_035396 rs843704 |
109 | G>S | No |
UniProt dbSNP |
|
| TCGA novel | 113 | N>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV000946610 rs13424808 VAR_035397 |
116 | P>L | No |
ClinVar UniProt dbSNP |
|
| TCGA novel | 121 | H>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs771327631 | 122 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1306275078 | 138 | E>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762227245 | 169 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs371422603 | 173 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs973521434 | 174 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 175 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1309269585 | 221 | R>G | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 240 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
VAR_035398 rs17189743 |
246 | R>C | No |
UniProt dbSNP |
|
| TCGA novel | 255 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 303 | Y>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1165531606 | 307 | K>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1410110511 | 331 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773922991 | 331 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs531572033 | 333 | G>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 335 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 337 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs371089329 | 344 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 348 | C>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1465893390 | 356 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 361 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1453563318 | 367 | Q>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 371 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757162814 | 392 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1423931033 | 399 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 403 | R>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 404 | P>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with Q8N831
No regional properties for Q8N831
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q8N831 | |||
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| histone binding | Binding to a histone, any of a group of water-soluble proteins found in association with the DNA of eukaryotic or archaeal chromosomes. They are involved in the condensation and coiling of chromosomes during cell division and have also been implicated in gene regulation and DNA replication. They may be chemically modified (methylated, acetlyated and others) to regulate gene transcription. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| nucleosome assembly | The aggregation, arrangement and bonding together of a nucleosome, the beadlike structural units of eukaryotic chromatin composed of histones and DNA. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O19110 | TSPY1 | Testis-specific Y-encoded protein 1 | Bos taurus (Bovine) | PR |
| P0DME0 | SETSIP | Protein SETSIP | Homo sapiens (Human) | SS |
| Q9H2G4 | TSPYL2 | Testis-specific Y-encoded-like protein 2 | Homo sapiens (Human) | PR |
| Q99457 | NAP1L3 | Nucleosome assembly protein 1-like 3 | Homo sapiens (Human) | PR |
| P55209 | NAP1L1 | Nucleosome assembly protein 1-like 1 | Homo sapiens (Human) | PR |
| Q01534 | TSPY1 | Testis-specific Y-encoded protein 1 | Homo sapiens (Human) | PR |
| P0CV99 | TSPY4 | Testis-specific Y-encoded protein 4 | Homo sapiens (Human) | PR |
| P0CV98 | TSPY3 | Testis-specific Y-encoded protein 3 | Homo sapiens (Human) | PR |
| P0CW00 | TSPY8 | Testis-specific Y-encoded protein 8 | Homo sapiens (Human) | PR |
| P0CW01 | TSPY10 | Testis-specific Y-encoded protein 10 | Homo sapiens (Human) | PR |
| A6NKD2 | TSPY2 | Testis-specific Y-encoded protein 2 | Homo sapiens (Human) | PR |
| Q01105 | SET | Protein SET | Homo sapiens (Human) | SS |
| Q9H489 | TSPY26P | Putative testis-specific Y-encoded-like protein 3 | Homo sapiens (Human) | PR |
| Q9EQU5 | Set | Protein SET | Mus musculus (Mouse) | EV |
| Q63945 | Set | Protein SET | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSLPESPHSP | ATLDYALEDP | HQGQRSREKS | KATEVMADMF | DGRLEPIVFP | PPRLPEEGVA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PQDPADGGHT | FHILVDAGRS | HGAIKAGQEV | TPPPAEGLEA | ASASLTTDGS | LKNGFPGEET |
| 130 | 140 | 150 | 160 | 170 | 180 |
| HGLGGEKALE | TCGAGRSESE | VIAEGKAEDV | KPEECAMFSA | PVDEKPGGEE | MDVAEENRAI |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DEVNREAGPG | PGPGPLNVGL | HLNPLESIQL | ELDSVNAEAD | RALLQVERRF | GQIHEYYLEQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RNDIIRNIPG | FWVTAFRHHP | QLSAMIRGQD | AEMLSYLTNL | EVKELRHPRT | GCKFKFFFQR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NPYFRNKLIV | KVYEVRSFGQ | VVSFSTLIMW | RRGHGPQSFI | HRNRHVICSF | FTWFSDHSLP |
| 370 | 380 | 390 | 400 | ||
| ESDRIAQIIK | EDLWSNPLQY | YLLGEDAHRA | RRRLVREPVE | IPRPFGFQCG |