Q8N1B3
Gene name |
CCNQ |
Protein name |
Cyclin-Q |
Names |
CDK10-activating cyclin, Cyclin-M, Cyclin-related protein FAM58A |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:92002 |
EC number |
|
Protein Class |
|
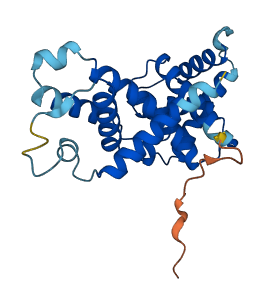
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q8N1B3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8N1B3-F1 | Predicted | AlphaFoldDB |
179 variants for Q8N1B3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000011420 rs1569536891 |
102 | N>* | Syndactyly-telecanthus-anogenital and renal malformations syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
CA337228376 rs782010600 |
2 | E>D | No |
ClinGen ExAC |
|
|
rs782127205 CA337228379 |
2 | E>G | No |
ClinGen ExAC |
|
|
CA415091973 rs1461603323 |
3 | A>V | No |
ClinGen TOPMed |
|
|
CA415091960 rs1557027849 |
4 | P>L | No |
ClinGen gnomAD |
|
|
CA415091946 rs1557027846 |
5 | E>G | No |
ClinGen gnomAD |
|
|
rs200645137 RCV000869483 |
6 | G>missing | No |
ClinVar dbSNP |
|
|
CA415091919 rs868990609 |
7 | G>R | No |
ClinGen Ensembl |
|
|
CA415091902 rs1557027829 |
8 | G>* | No |
ClinGen gnomAD |
|
| TCGA novel | 9 | G>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 9 | G>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557027823 CA415091879 |
9 | G>V | No |
ClinGen gnomAD |
|
|
rs781939131 CA337228324 |
10 | G>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA337228316 rs782235940 |
11 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs782235940 CA415091858 |
11 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1557027804 CA415091841 |
12 | A>T | No |
ClinGen gnomAD |
|
|
rs1557027803 CA415091830 |
12 | A>V | No |
ClinGen gnomAD |
|
|
rs1557027796 CA415091819 |
13 | A>E | No |
ClinGen gnomAD |
|
|
rs782392509 CA337228304 |
13 | A>S | No |
ClinGen ExAC |
|
|
rs1557027790 CA415091805 |
14 | R>Q | No |
ClinGen gnomAD |
|
|
CA415091790 rs1557027789 |
15 | G>A | No |
ClinGen gnomAD |
|
|
CA415091792 rs1557027789 |
15 | G>V | No |
ClinGen gnomAD |
|
|
rs782278351 CA415091771 |
16 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA337228302 rs782278351 |
16 | P>Q | No |
ClinGen ExAC gnomAD |
|
|
CA337228296 rs200171156 |
17 | E>G | No |
ClinGen gnomAD |
|
|
CA337228277 rs782681960 |
19 | Q>E | No |
ClinGen ExAC |
|
|
rs1557027772 CA415091722 |
19 | Q>H | No |
ClinGen gnomAD |
|
|
CA415091704 rs1446347435 |
20 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1446347435 CA415091708 |
20 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs1557027764 CA415091712 |
20 | P>S | No |
ClinGen gnomAD |
|
|
rs1557027764 CA415091717 |
20 | P>T | No |
ClinGen gnomAD |
|
|
rs782316470 CA337228272 |
21 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA415091696 rs1557027757 |
21 | A>S | No |
ClinGen gnomAD |
|
|
rs1557027757 CA415091701 |
21 | A>T | No |
ClinGen gnomAD |
|
|
rs782316470 CA415091693 |
21 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA415091673 rs1557027741 |
22 | P>H | No |
ClinGen gnomAD |
|
|
rs1557027735 CA415091670 |
23 | E>* | No |
ClinGen gnomAD |
|
|
CA415091660 rs1557027729 |
24 | A>D | No |
ClinGen gnomAD |
|
|
CA415091662 rs1557027732 |
24 | A>S | No |
ClinGen gnomAD |
|
|
CA415091652 rs1235293786 |
25 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1557027720 CA415091649 |
26 | V>E | No |
ClinGen gnomAD |
|
|
CA337228263 rs782611841 |
26 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA415091619 rs1209434931 |
28 | F>Y | No |
ClinGen TOPMed |
|
|
rs1284916925 CA415091602 |
29 | R>Q | No |
ClinGen TOPMed |
|
|
rs1603165965 CA415091594 |
30 | V>M | No |
ClinGen Ensembl |
|
|
rs1557027711 CA415091580 |
31 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 31 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557027708 CA415091553 |
32 | R>S | No |
ClinGen gnomAD |
|
|
rs1448636185 CA415091504 |
35 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
CA415091501 rs1185167911 |
35 | M>T | No |
ClinGen TOPMed |
|
|
CA337227250 rs781881866 |
40 | K>R | No |
ClinGen Ensembl |
|
|
CA10548480 rs537796562 |
43 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA415090708 rs1400512637 |
44 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
CA415090712 rs1400512637 |
44 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs781948889 CA10548479 |
44 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 46 | I>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA415090661 rs1557027054 |
48 | I>V | No |
ClinGen gnomAD |
|
|
CA337227228 rs982875664 |
49 | A>V | No |
ClinGen Ensembl |
|
| TCGA novel | 54 | I>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 56 | H>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10548475 rs782646629 |
56 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1557027043 CA415090509 |
58 | F>L | No |
ClinGen gnomAD |
|
|
CA10548473 rs782293018 |
60 | C>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 60 | C>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1603162377 CA415090487 |
61 | E>D | No |
ClinGen Ensembl |
|
|
rs149734965 RCV000894388 CA10548471 |
61 | E>K | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1557027031 CA415090476 |
63 | N>S | No |
ClinGen gnomAD |
|
|
CA10548470 rs376670332 |
64 | L>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA415090459 rs1603162343 |
66 | A>P | No |
ClinGen Ensembl |
|
|
CA10548467 rs781987078 RCV000996038 |
67 | Y>C | No |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
rs1023545906 CA337227142 |
67 | Y>H | No |
ClinGen Ensembl |
|
| TCGA novel | 72 | I>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA415090347 rs1557027008 |
75 | S>A | No |
ClinGen gnomAD |
|
|
rs782228548 CA10548465 |
75 | S>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
rs781801497 CA10548464 |
78 | Y>D | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 80 | A>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs781921854 CA10548463 |
81 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs1355426525 CA415090245 |
83 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
CA10548461 rs781977080 |
86 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782390185 CA10548460 |
87 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA415090183 rs1557026982 |
87 | H>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 89 | R>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557026970 CA415090168 |
89 | R>Q | No |
ClinGen gnomAD |
|
|
CA415090171 rs1442543413 |
89 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
rs782147876 CA10548459 |
90 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs1188961308 CA415090148 |
91 | R>C | No |
ClinGen TOPMed |
|
|
CA415090123 rs1557026955 |
93 | I>V | No |
ClinGen gnomAD |
|
|
CA10548458 rs782031283 |
95 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 98 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10548457 rs188044074 |
98 | N>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA415089942 rs1557026614 |
99 | R>S | No |
ClinGen gnomAD |
|
|
CA415089903 rs1557026607 |
102 | N>D | No |
ClinGen gnomAD |
|
| TCGA novel | 105 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10548431 rs782206940 |
105 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs782608728 CA10548430 |
107 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA415089817 rs1397964100 |
109 | E>Q | No |
ClinGen TOPMed |
|
|
rs782498226 CA10548429 |
112 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs201043377 CA10548428 |
113 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
RCV000872593 rs150562029 RCV002227501 CA10548427 |
113 | R>H | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs782558422 CA10548426 |
115 | W>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1273846093 CA415089710 |
117 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs918989098 CA337226278 |
118 | R>G | No |
ClinGen gnomAD |
|
|
rs918989098 CA415089697 |
118 | R>W | No |
ClinGen gnomAD |
|
|
rs782469736 CA10548423 |
121 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA415089655 rs1345170998 |
121 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA337226266 rs981165355 |
122 | V>A | No |
ClinGen Ensembl |
|
|
rs1557026564 CA415089644 |
122 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA10548422 rs781843718 |
128 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs183758563 CA10548421 |
129 | L>R | No |
ClinGen 1000Genomes ExAC |
|
|
rs1199674666 CA415089502 |
132 | L>V | No |
ClinGen TOPMed |
|
|
CA10548420 rs781799914 |
133 | R>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1240480716 CA415089488 |
133 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA415089402 rs1557026547 |
139 | Q>R | No |
ClinGen gnomAD |
|
|
rs1557026545 CA415089377 |
141 | P>A | No |
ClinGen gnomAD |
|
| TCGA novel | 141 | P>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs374423745 CA10548419 |
142 | H>Y | No |
ClinGen ESP ExAC gnomAD |
|
| TCGA novel | 143 | K>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA415088623 rs1557026107 |
144 | Y>H | No |
ClinGen gnomAD |
|
|
CA415088352 rs1557026099 |
156 | L>M | No |
ClinGen gnomAD |
|
|
rs781795178 CA10548401 |
158 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1557026096 CA415088305 |
158 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA10548400 rs782734343 |
163 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA415088193 rs782734343 |
163 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1393228949 CA415088196 |
163 | R>W | No |
ClinGen TOPMed |
|
|
CA337225364 rs372939037 |
164 | T>S | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA10548396 rs370810560 |
168 | V>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA10548397 rs370810560 |
168 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA415088093 rs1178567203 |
170 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1557026076 CA415088074 |
171 | W>* | No |
ClinGen gnomAD |
|
|
CA337225336 rs930788533 |
175 | R>Q | No |
ClinGen Ensembl |
|
|
CA10548394 rs782309279 |
175 | R>W | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 176 | D>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs781948832 CA10548392 |
177 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs1557026069 RCV000522853 CA415087938 |
180 | G>R | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs782408272 CA10548388 |
181 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782408272 CA10548389 |
181 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10548387 rs782298309 |
182 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA415087872 rs1557026054 |
183 | C>* | No |
ClinGen gnomAD |
|
|
CA337225278 VAR_034642 rs17850173 |
183 | C>S | No |
ClinGen UniProt Ensembl dbSNP |
|
| TCGA novel | 185 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA415087843 rs1557026048 |
185 | R>H | No |
ClinGen gnomAD |
|
|
CA337225271 rs191902079 |
185 | R>S | No |
ClinGen 1000Genomes |
|
|
CA415087826 rs1557026045 |
186 | F>Y | No |
ClinGen gnomAD |
|
|
rs1603157216 CA415087812 |
187 | Q>K | No |
ClinGen Ensembl |
|
| TCGA novel | 188 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 189 | Q>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA415087707 rs1557026041 |
191 | I>V | No |
ClinGen gnomAD |
|
|
rs1432027234 CA415087689 |
192 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1557026036 CA415087663 |
193 | V>L | No |
ClinGen gnomAD |
|
|
CA415087668 rs1557026036 |
193 | V>M | No |
ClinGen gnomAD |
|
|
CA415087644 rs1427113628 |
194 | A>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA10548383 rs782618270 |
195 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA415087578 rs1463781917 |
197 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
CA415087580 rs1463781917 |
197 | Y>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1557026021 CA415087498 |
202 | V>A | No |
ClinGen gnomAD |
|
|
CA10548381 rs781880341 |
202 | V>F | No |
ClinGen ExAC gnomAD |
|
|
rs1057521251 RCV000434465 CA16609136 |
206 | E>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA415087375 rs1557026001 |
209 | A>S | No |
ClinGen gnomAD |
|
|
CA415087378 rs1557026001 |
209 | A>T | No |
ClinGen gnomAD |
|
|
CA10548377 rs782742456 |
210 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs146203325 CA337225224 |
212 | E>K | No |
ClinGen ESP TOPMed |
|
| TCGA novel | 213 | A>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10548374 rs782770419 |
216 | P>L | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 216 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV001200550 rs2090998416 |
219 | Q>* | No |
ClinVar dbSNP |
|
|
rs782038624 CA10548372 |
219 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
CA415085887 rs1557025069 |
221 | F>L | No |
ClinGen gnomAD |
|
|
rs1557025062 CA415085787 |
224 | D>N | No |
ClinGen gnomAD |
|
|
CA337223689 rs1025698292 |
228 | P>A | No |
ClinGen TOPMed |
|
|
rs782739507 CA10548358 |
232 | N>K | No |
ClinGen ExAC |
|
| TCGA novel | 233 | I>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA415085461 rs782481531 |
237 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10548357 rs782481531 |
237 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs112616337 CA337223669 |
238 | I>M | No |
ClinGen Ensembl |
|
|
rs141657773 CA10548356 RCV000870882 |
242 | T>A | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA415085382 rs1557025046 |
243 | M>V | No |
ClinGen gnomAD |
|
|
rs1557025044 CA415085363 |
244 | D>G | No |
ClinGen gnomAD |
|
|
rs868934018 CA415085337 |
246 | E>G | No |
ClinGen TOPMed |
|
|
rs782304286 CA10548355 |
246 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
1 associated diseases with Q8N1B3
[MIM: 300707]: Toe syndactyly, telecanthus, and anogenital and renal malformations (STAR)
A syndrome characterized by anal, genital and renal tract anomalies, facial dysmorphism and syndactyly. Features include anal stenosis, a rectovaginal fistula, clitoral hypertrophy, a pelvic right kidney, toe syndactyly, and telecanthus. {ECO:0000269|PubMed:18297069}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A syndrome characterized by anal, genital and renal tract anomalies, facial dysmorphism and syndactyly. Features include anal stenosis, a rectovaginal fistula, clitoral hypertrophy, a pelvic right kidney, toe syndactyly, and telecanthus. {ECO:0000269|PubMed:18297069}. Note=The disease is caused by variants affecting the gene represented in this entry.
3 regional properties for Q8N1B3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Cyclin, N-terminal | 29 - 136 | IPR006671 |
| domain | Cyclin-like domain | 32 - 130 | IPR013763-1 |
| domain | Cyclin-like domain | 150 - 239 | IPR013763-2 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cyclin-dependent protein kinase holoenzyme complex | Cyclin-dependent protein kinases (CDKs) are enzyme complexes that contain a kinase catalytic subunit associated with a regulatory cyclin partner. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| cyclin-dependent protein serine/threonine kinase regulator activity | Modulates the activity of a cyclin-dependent protein serine/threonine kinase, enzymes of the protein kinase family that are regulated through association with cyclins and other proteins. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| regulation of cell cycle G2/M phase transition | Any signalling pathway that modulates the activity of a cell cycle cyclin-dependent protein kinase to modulate the switch from G2 phase to M phase of the cell cycle. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q6T8E9 | CCNT1 | Cyclin-T1 | Bos taurus (Bovine) | SS |
| Q8HXN7 | CCNT1 | Cyclin-T1 | Pan troglodytes (Chimpanzee) | SS |
| P24863 | CCNC | Cyclin-C | Homo sapiens (Human) | PR |
| P51946 | CCNH | Cyclin-H | Homo sapiens (Human) | PR |
| O60563 | CCNT1 | Cyclin-T1 | Homo sapiens (Human) | EV |
| O75909 | CCNK | Cyclin-K | Homo sapiens (Human) | PR |
| O88874 | Ccnk | Cyclin-K | Mus musculus (Mouse) | PR |
| Q9QWV9 | Ccnt1 | Cyclin-T1 | Mus musculus (Mouse) | SS |
| Q6Z7H3 | CYCT1_2 | Cyclin-T1-2 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAC5 | CYCT1-3 | Cyclin-T1-3 | Oryza sativa subsp japonica (Rice) | PR |
| Q2QQS5 | CYCT1-1 | Cyclin-T1-4 | Oryza sativa subsp japonica (Rice) | PR |
| Q8GYM6 | CYCT1-4 | Cyclin-T1-4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q503D6 | ccnq | Cyclin-Q | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEAPEGGGGG | PAARGPEGQP | APEARVHFRV | ARFIMEAGVK | LGMRSIPIAT | ACTIYHKFFC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ETNLDAYDPY | LIAMSSIYLA | GKVEEQHLRT | RDIINVSNRY | FNPSGEPLEL | DSRFWELRDS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IVQCELLMLR | VLRFQVSFQH | PHKYLLHYLV | SLQNWLNRHS | WQRTPVAVTA | WALLRDSYHG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ALCLRFQAQH | IAVAVLYLAL | QVYGVEVPAE | VEAEKPWWQV | FNDDLTKPII | DNIVSDLIQI |
| YTMDTEIP |