Q8LST2
Gene name |
WNK7 (At1g49160, F27J15.7) |
Protein name |
Probable serine/threonine-protein kinase WNK7 |
Names |
AtWNK7, Protein kinase with no lysine 7 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G49160 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
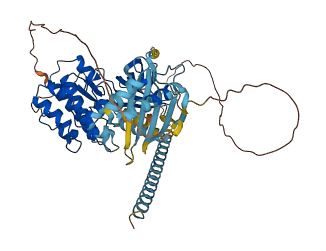
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
174-195 (Activation loop from InterPro)
Target domain |
28-285 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q8LST2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8LST2-F1 | Predicted | AlphaFoldDB |
40 variants for Q8LST2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH01326534 | 4 | S>P | No | 1000Genomes | |
| tmp_1_18181845_G_C | 8 | S>C | No | 1000Genomes | |
| tmp_1_18181842_G_A | 9 | A>V | No | 1000Genomes | |
| ENSVATH04912033 | 29 | R>W | No | 1000Genomes | |
| tmp_1_18181359_G_A | 80 | L>F | No | 1000Genomes | |
| ENSVATH04912022 | 101 | N>S | No | 1000Genomes | |
| tmp_1_18181278_T_C | 107 | I>V | No | 1000Genomes | |
| ENSVATH04912019 | 135 | A>G | No | 1000Genomes | |
| tmp_1_18181090_C_T | 136 | R>K | No | 1000Genomes | |
| tmp_1_18180945_T_A | 184 | Q>H | No | 1000Genomes | |
| tmp_1_18180750_C_T | 217 | M>I | No | 1000Genomes | |
| ENSVATH04912013 | 226 | E>D | No | 1000Genomes | |
| ENSVATH04912009 | 251 | L>V | No | 1000Genomes | |
| tmp_1_18180403_G_A | 295 | P>S | No | 1000Genomes | |
| tmp_1_18180390_G_A | 299 | P>L | No | 1000Genomes | |
| tmp_1_18180375_G_A | 304 | P>L | No | 1000Genomes | |
| tmp_1_18180333_C_T | 318 | G>E | No | 1000Genomes | |
| tmp_1_18180328_G_A | 320 | P>S | No | 1000Genomes | |
| tmp_1_18180297_T_G | 330 | N>T | No | 1000Genomes | |
| tmp_1_18180292_C_T | 332 | D>N | No | 1000Genomes | |
| ENSVATH04912006 | 343 | S>R | No | 1000Genomes | |
| ENSVATH13458123 | 347 | G>E | No | 1000Genomes | |
| ENSVATH00089040 | 349 | N>K | No | 1000Genomes | |
| tmp_1_18180206_G_T | 360 | N>K | No | 1000Genomes | |
| ENSVATH14267537 | 385 | G>E | No | 1000Genomes | |
| ENSVATH01326520 | 403 | N>T | No | 1000Genomes | |
| ENSVATH14267536 | 442 | A>T | No | 1000Genomes | |
| tmp_1_18179765_T_A | 461 | N>I | No | 1000Genomes | |
| tmp_1_18179763_C_A | 462 | G>C | No | 1000Genomes | |
| tmp_1_18179742_C_T | 469 | E>K | No | 1000Genomes | |
| tmp_1_18179708_C_T | 480 | C>Y | No | 1000Genomes | |
| tmp_1_18179703_G_C | 482 | R>G | No | 1000Genomes | |
| ENSVATH04912002 | 491 | V>L | No | 1000Genomes | |
| tmp_1_18179666_G_T | 494 | T>K | No | 1000Genomes | |
| ENSVATH04912001 | 503 | P>L | No | 1000Genomes | |
| ENSVATH01326519 | 513 | E>D | No | 1000Genomes | |
| ENSVATH04912000 | 516 | K>T | No | 1000Genomes | |
| ENSVATH14267535 | 541 | T>K | No | 1000Genomes | |
| tmp_1_18179503_A_T | 548 | F>L | No | 1000Genomes | |
| tmp_1_18179499_C_G | 550 | E>Q | No | 1000Genomes |
No associated diseases with Q8LST2
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
19 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q96J92 | WNK4 | Serine/threonine-protein kinase WNK4 | Homo sapiens (Human) | SS |
| Q9BYP7 | WNK3 | Serine/threonine-protein kinase WNK3 | Homo sapiens (Human) | SS |
| Q9H4A3 | WNK1 | Serine/threonine-protein kinase WNK1 | Homo sapiens (Human) | SS |
| Q9Y3S1 | WNK2 | Serine/threonine-protein kinase WNK2 | Homo sapiens (Human) | SS |
| Q9UHY1 | NRBP1 | Nuclear receptor-binding protein | Homo sapiens (Human) | PR |
| P83741 | Wnk1 | Serine/threonine-protein kinase WNK1 | Mus musculus (Mouse) | SS |
| Q3UH66 | Wnk2 | Serine/threonine-protein kinase WNK2 | Mus musculus (Mouse) | SS |
| Q80UE6 | Wnk4 | Serine/threonine-protein kinase WNK4 | Mus musculus (Mouse) | SS |
| Q80XP9 | Wnk3 | Serine/threonine-protein kinase WNK3 | Mus musculus (Mouse) | SS |
| Q99J45 | Nrbp1 | Nuclear receptor-binding protein | Mus musculus (Mouse) | PR |
| Q6R2V0 | WNK1 | Serine/threonine-protein kinase WNK1 | Sus scrofa (Pig) | SS |
| Q7TPK6 | Wnk4 | Serine/threonine-protein kinase WNK4 | Rattus norvegicus (Rat) | SS |
| Q9JIH7 | Wnk1 | Serine/threonine-protein kinase WNK1 | Rattus norvegicus (Rat) | EV |
| E9PTG8 | Stk10 | Serine/threonine-protein kinase 10 | Rattus norvegicus (Rat) | PR |
| X5M5N0 | wnk-1 | Serine/threonine-protein kinase WNK | Caenorhabditis elegans | SS |
| Q8RXE5 | WNK10 | Probable serine/threonine-protein kinase WNK10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q944Q0 | WNK8 | Serine/threonine-protein kinase WNK8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LVL5 | WNK4 | Probable serine/threonine-protein kinase WNK4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STK6 | WNK3 | Probable serine/threonine-protein kinase WNK3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEGSEDASAI | VEPPDPEVLE | IDPTCRYIRY | KEVIGKGASK | TVFKGFDEVD | GIEVAWNQVR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IDDLLQSPDC | LERLYSEVRL | LKSLKHKNII | RFYNSWIDDK | NKTVNIITEL | FTSGSLRQYR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KKHRKVNMKA | VKCWARQILT | GLKYLHSQDP | PIIHRDIKCD | NIFINGNHGE | VKIGDLGLAT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VMEQANAKSV | IGTPEFMAPE | LYDENYNELA | DIYSFGMCML | EMVTFEYPYC | ECRNSAQIYK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KVSSGIKPAS | LSKVKDPEVM | KFIEKCLLPA | SERLSAEELL | LDSFLNVNGL | VMNNPLPLPD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| IVMPKEGSFG | ERCLMSEGPP | NARNRTMSMN | LDEDNNLPIV | ISSNNSGTNC | IEVRRAKRGN |
| 370 | 380 | 390 | 400 | 410 | 420 |
| FFVLKGEEND | ENSVSLILRI | VDENGRVRNI | HFLFFQEGDT | ASNVSSEMVE | QLELTDKNVK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| FIAELIDVLL | VNLIPNWKTD | VAVDHLIHPQ | QNQSSKDNHQ | NGASSQAGES | ISHSLSSDYC |
| 490 | 500 | 510 | 520 | 530 | 540 |
| PRSDDEANPT | VAATTEDQEA | EKPGSLEEEE | EDERLKEELE | KIEERFREEM | KEITRKREEA |
| 550 | |||||
| TMETKNRFFE | KKMQQVE |