Q8K4T3
Gene name |
Stradb (Als2cr2, Papk, Syradb) |
Protein name |
STE20-related kinase adapter protein beta |
Names |
STRAD beta, Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 2 protein homolog, ILP-interacting protein homolog, Polyploidy-associated protein kinase, Pseudokinase ALS2CR2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:227154 |
EC number |
|
Protein Class |
|
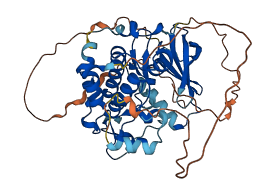
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q8K4T3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8K4T3-F1 | Predicted | AlphaFoldDB |
32 variants for Q8K4T3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs255507662 | 49 | V>A | No | EVA | |
| rs228938701 | 54 | D>N | No | EVA | |
| rs3388471170 | 59 | E>G | No | EVA | |
| rs3388471441 | 71 | L>* | No | EVA | |
| rs3388471425 | 79 | H>Y | No | EVA | |
| rs3390046161 | 92 | N>Y | No | EVA | |
| rs3390036263 | 94 | E>* | No | EVA | |
| rs3389980527 | 102 | K>Q | No | EVA | |
| rs3388471121 | 116 | H>D | No | EVA | |
| rs3388472571 | 133 | W>* | No | EVA | |
| rs3388472045 | 136 | S>Y | No | EVA | |
| rs3388472432 | 146 | Q>K | No | EVA | |
| rs3388473416 | 158 | E>V | No | EVA | |
| rs3388473363 | 164 | I>S | No | EVA | |
| rs3388472943 | 180 | C>* | No | EVA | |
| rs3388470546 | 205 | H>L | No | EVA | |
| rs3388472439 | 215 | R>M | No | EVA | |
| rs3411306025 | 219 | V>M | No | EVA | |
| rs3390015301 | 226 | S>C | No | EVA | |
| rs3388472869 | 235 | P>S | No | EVA | |
| rs3411152980 | 260 | E>* | No | EVA | |
| rs3388472301 | 296 | Q>L | No | EVA | |
| rs3388471965 | 347 | Q>* | No | EVA | |
| rs3388471791 | 367 | V>I | No | EVA | |
| rs3388472601 | 368 | F>L | No | EVA | |
| rs1131736529 | 374 | E>A | No | EVA | |
| rs1134385358 | 375 | E>A | No | EVA | |
| rs1134718319 | 377 | Q>K | No | EVA | |
| rs1133212700 | 379 | S>F | No | EVA | |
| rs247646675 | 396 | Q>P | No | EVA | |
| rs3388470527 | 398 | V>L | No | EVA | |
| rs3388474187 | 403 | E>K | No | EVA |
No associated diseases with Q8K4T3
1 regional properties for Q8K4T3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 58 - 369 | IPR000719 |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| aggresome | An inclusion body formed by dynein-dependent retrograde transport of an aggregated protein on microtubules. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| serine/threonine protein kinase complex | A protein complex which is capable of protein serine/threonine kinase activity. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine/threonine kinase activator activity | Binds to and increases the activity of a protein serine/threonine kinase. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| activation of protein kinase activity | Any process that initiates the activity of an inactive protein kinase. |
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| cell morphogenesis | The developmental process in which the size or shape of a cell is generated and organized. |
| cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures. |
| JNK cascade | An intracellular protein kinase cascade containing at least a JNK (a MAPK), a JNKK (a MAPKK) and a JUN3K (a MAP3K). The cascade can also contain an additional tier: the upstream MAP4K. The kinases in each tier phosphorylate and activate the kinases in the downstream tier to transmit a signal within a cell. |
| negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | Any process that stops, prevents or reduces the frequency, rate or extent of extrinsic apoptotic signaling pathway in absence of ligand. |
| protein export from nucleus | The directed movement of a protein from the nucleus into the cytoplasm. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O95747 | OXSR1 | Serine/threonine-protein kinase OSR1 | Homo sapiens (Human) | PR |
| Q9UEW8 | STK39 | STE20/SPS1-related proline-alanine-rich protein kinase | Homo sapiens (Human) | PR |
| Q7RTN6 | STRADA | STE20-related kinase adapter protein alpha | Homo sapiens (Human) | PR |
| Q9C0K7 | STRADB | STE20-related kinase adapter protein beta | Homo sapiens (Human) | PR |
| Q6P9R2 | Oxsr1 | Serine/threonine-protein kinase OSR1 | Mus musculus (Mouse) | PR |
| Q9Z1W9 | Stk39 | STE20/SPS1-related proline-alanine-rich protein kinase | Mus musculus (Mouse) | PR |
| Q3UUJ4 | Strada | STE20-related kinase adapter protein alpha | Mus musculus (Mouse) | PR |
| Q863I2 | OXSR1 | Serine/threonine-protein kinase OSR1 | Sus scrofa (Pig) | PR |
| Q7TNZ6 | Strada | STE20-related kinase adapter protein alpha | Rattus norvegicus (Rat) | PR |
| O88506 | Stk39 | STE20/SPS1-related proline-alanine-rich protein kinase | Rattus norvegicus (Rat) | PR |
| O23304 | BLUS1 | Serine/threonine-protein kinase BLUS1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSLLDCFCAS | RTRVESLRPE | KQSETSIHQY | LVDESAISRP | PPSARASEVI | CSTDVSHYEL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QVEIGRGFDN | LTSVHLARHT | PTGTLVTVKI | TNLESCTEER | LKALQRAVIL | SHFFQHPNIT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TYWTVFTVGS | WLWVISPFMA | YGSASQLLRT | YFPDGMSETL | IRNILFGAVQ | GLNYLHQNGC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IHRSFKASHI | LISGDGLVTL | SGLSHLHSLL | KHGQRHRAVF | DFPQFSTSVQ | PWLSPELLRQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DLHGYNVKSD | IYSVGITACE | LASGQVPFQD | MHRTQMLLQK | LKGPPYSPLD | VSIFPQSDSR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MRNSQSGVDS | GIGESVLVST | GTHTVNSDRL | HTPSTKTFSP | AFFSLVQLCL | QQDPEKRPSA |
| 370 | 380 | 390 | 400 | 410 | |
| SSLLSHVFFK | QMKEESQDSI | LPLLPPAYNR | PSASLQPVSP | WSELEFQFPD | DKDPVWEF |