Q8IYK8
Gene name |
REM2 |
Protein name |
GTP-binding protein REM 2 |
Names |
Rad and Gem-like GTP-binding protein 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:161253 |
EC number |
|
Protein Class |
|
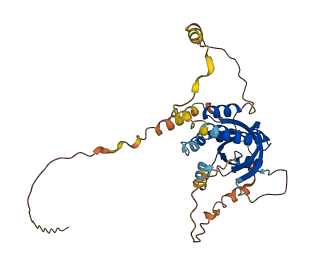
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q8IYK8
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3CBQ | X-ray | 182 A | A | 110-286 | PDB |
| AF-Q8IYK8-F1 | Predicted | AlphaFoldDB |
265 variants for Q8IYK8
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA388937187 rs1286284219 |
3 | T>M | No |
ClinGen gnomAD |
|
|
CA388937203 rs1230815173 |
6 | D>N | No |
ClinGen TOPMed |
|
|
rs377287826 CA7106118 |
9 | M>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1218135218 CA388937243 |
11 | M>T | No |
ClinGen gnomAD |
|
|
rs1487898657 CA388937254 |
12 | D>E | No |
ClinGen gnomAD |
|
|
CA388937251 rs1283825832 |
12 | D>V | No |
ClinGen gnomAD |
|
|
rs1297229473 CA388937295 |
19 | C>R | No |
ClinGen TOPMed |
|
|
CA388937307 rs1181561519 |
20 | P>L | No |
ClinGen gnomAD |
|
|
CA388937304 rs1471678477 |
20 | P>S | No |
ClinGen gnomAD |
|
| TCGA novel | 21 | S>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA388937319 rs1441679922 |
22 | G>V | No |
ClinGen gnomAD |
|
|
rs1360487820 CA388937323 |
23 | S>N | No |
ClinGen gnomAD |
|
|
rs965656400 CA257734570 |
24 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA388937330 rs1357847466 |
24 | R>H | No |
ClinGen gnomAD |
|
|
CA7106124 rs368184584 |
25 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7106125 rs368184584 |
25 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs374573401 CA7106123 |
25 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs777061887 CA7106126 |
27 | S>A | No |
ClinGen ExAC |
|
|
rs1429908005 CA388937346 |
28 | P>T | No |
ClinGen TOPMed |
|
|
rs1352910787 CA388937356 |
29 | P>L | No |
ClinGen gnomAD |
|
|
CA257734610 rs373808086 |
30 | G>E | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs151172050 CA7106127 |
30 | G>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA388937363 rs1440524193 |
31 | T>A | No |
ClinGen gnomAD |
|
|
rs201949801 CA388937365 |
31 | T>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs201949801 CA7106128 |
31 | T>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA388937367 rs1370594181 |
32 | P>T | No |
ClinGen TOPMed |
|
|
CA388937380 rs1176863333 |
34 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA388937410 rs1477947960 |
36 | A>G | No |
ClinGen gnomAD |
|
| TCGA novel | 36 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7106137 rs778534880 |
37 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1218942988 CA388937423 |
38 | A>V | No |
ClinGen gnomAD |
|
|
CA7106139 rs201532289 |
39 | T>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1405468374 CA388937438 |
41 | L>Q | No |
ClinGen gnomAD |
|
|
CA7106143 rs776972071 |
43 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA388937461 rs1435603312 |
45 | E>Q | No |
ClinGen gnomAD |
|
|
CA388937472 rs1176503849 |
46 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA7106145 rs748689778 |
47 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA7106146 rs372126836 |
52 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7106148 rs555231261 |
53 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA7106147 rs773368193 |
53 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs376129701 CA7106151 |
55 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA388937540 rs1173262758 |
57 | P>S | No |
ClinGen TOPMed |
|
|
CA388937555 rs1265933500 |
59 | A>V | No |
ClinGen gnomAD |
|
|
rs369377234 CA7106153 |
61 | G>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs369377234 CA7106152 |
61 | G>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1566498862 CA388937575 |
63 | P>H | No |
ClinGen Ensembl |
|
|
rs1372180106 CA388937578 |
64 | R>* | No |
ClinGen gnomAD |
|
| TCGA novel | 64 | R>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1184522785 CA388937581 |
64 | R>I | No |
ClinGen gnomAD |
|
|
rs1412960896 CA388937584 |
64 | R>S | No |
ClinGen gnomAD |
|
|
CA7106154 rs200631469 |
65 | R>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs894910280 CA257735086 |
65 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA388937590 rs1164509930 |
66 | R>K | No |
ClinGen gnomAD |
|
|
rs765044356 CA7106155 |
67 | G>C | No |
ClinGen ExAC gnomAD |
|
|
rs1277260652 CA388937623 |
71 | V>I | No |
ClinGen gnomAD |
|
|
rs1247326010 CA388937631 |
72 | P>S | No |
ClinGen gnomAD |
|
|
CA7106159 rs757787473 |
73 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
CA388937640 rs1211610457 |
73 | Y>C | No |
ClinGen TOPMed |
|
| rs767618692 | 73 | Y>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA388937654 rs1355780384 |
75 | H>R | No |
ClinGen gnomAD |
|
|
CA388937668 rs1458777210 |
77 | L>F | No |
ClinGen gnomAD |
|
|
CA7106161 rs763818736 |
78 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs779523287 CA7106160 |
78 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs748416414 CA7106164 |
79 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7106163 rs61752841 |
79 | R>W | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7106165 rs770366952 |
81 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs761225229 CA7106166 |
81 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1366323174 CA388937695 |
82 | A>V | No |
ClinGen gnomAD |
|
|
CA7106168 CA7106169 rs771195142 |
84 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs759878174 CA7106170 |
88 | W>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388937749 rs1417718352 |
90 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA388937748 rs1417718352 |
90 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
CA257735115 rs907785013 |
91 | Q>* | No |
ClinGen TOPMed |
|
|
CA7106171 rs577629448 |
92 | A>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA7106172 rs577629448 |
92 | A>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 93 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA388937770 rs1397528245 |
94 | S>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA7106173 rs8014119 VAR_055938 |
96 | G>A | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1204230900 CA388937789 |
97 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA7106175 rs750382226 |
98 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA388937805 rs1311471515 |
100 | S>A | No |
ClinGen gnomAD |
|
|
CA388937809 rs1009519855 |
101 | L>M | No |
ClinGen gnomAD |
|
|
rs762941371 CA7106176 |
106 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA388937851 rs1566499006 |
107 | A>G | No |
ClinGen Ensembl |
|
| TCGA novel | 107 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1240135395 CA388937855 |
108 | P>S | No |
ClinGen gnomAD |
|
|
CA7106177 rs765909465 |
110 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA7106178 rs751044093 |
111 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1475208503 CA388937883 |
112 | D>G | No |
ClinGen TOPMed |
|
| TCGA novel | 115 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA7106180 rs780787465 |
115 | F>S | No |
ClinGen ExAC gnomAD |
|
|
CA7106182 rs199760801 |
118 | M>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7106184 rs373274304 |
121 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs377139889 CA7106185 |
122 | E>V | No |
ClinGen ESP ExAC TOPMed |
|
|
rs143212029 CA7106187 |
124 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA7106189 rs775515596 |
125 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs1356115824 CA388937973 |
126 | G>D | No |
ClinGen TOPMed |
|
|
CA7106190 rs760937195 |
128 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA388937995 rs200166181 |
129 | T>I | No |
ClinGen gnomAD |
|
|
rs200166181 CA257735148 |
129 | T>S | No |
ClinGen gnomAD |
|
|
CA7106191 rs769540596 |
131 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7106192 rs773026460 |
132 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA7106193 rs762922775 |
133 | T>N | No |
ClinGen ExAC gnomAD |
|
|
CA257735154 rs1049857366 |
134 | F>S | No |
ClinGen TOPMed |
|
|
rs1231575584 CA388938026 |
135 | G>D | No |
ClinGen gnomAD |
|
|
rs1375793198 CA388938031 |
136 | G>S | No |
ClinGen TOPMed |
|
|
rs1255710146 CA388938040 |
137 | L>R | No |
ClinGen gnomAD |
|
|
rs370177412 CA7106196 |
138 | Q>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA257735158 rs915188793 |
139 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA7106197 rs767173528 |
140 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752352297 CA7106198 |
141 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755788147 CA7106199 |
141 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA7106201 rs200710673 |
144 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA388938089 rs1465943474 |
145 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA7106202 rs186131311 |
145 | P>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA388938090 rs186131311 |
145 | P>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs186131311 CA7106203 |
145 | P>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA388938106 rs1176271055 |
147 | N>K | No |
ClinGen TOPMed |
|
|
rs1461568744 CA388938104 |
147 | N>S | No |
ClinGen gnomAD |
|
|
rs757637804 CA7106221 |
150 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751037447 CA388938152 |
152 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388938148 rs1595037891 |
152 | Y>D | No |
ClinGen Ensembl |
|
|
rs939579541 CA257735232 |
152 | Y>F | No |
ClinGen TOPMed |
|
|
rs1359444618 CA388938156 |
153 | E>G | No |
ClinGen gnomAD |
|
|
rs1056719322 CA257735236 |
153 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1173892297 CA388938166 |
154 | R>S | No |
ClinGen TOPMed |
|
|
rs769826661 CA7106226 |
155 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs769826661 CA7106225 |
155 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755189659 CA257735245 |
155 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7106227 rs755189659 |
155 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769826661 CA257735242 |
155 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388938171 rs1197309614 |
156 | I>N | No |
ClinGen TOPMed |
|
|
rs781412286 CA7106228 |
157 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388938176 rs1219595154 |
157 | M>V | No |
ClinGen gnomAD |
|
|
CA7106229 rs749097349 |
159 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs770774317 CA257735254 |
161 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7106230 rs770774317 |
161 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388938221 rs1566499373 |
163 | V>E | No |
ClinGen Ensembl |
|
|
CA388938231 rs1485417716 |
165 | L>V | No |
ClinGen gnomAD |
|
|
rs1487359286 CA388938241 |
167 | V>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
| TCGA novel | 168 | Y>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA257735260 rs772024367 |
169 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1416196249 CA388938257 |
169 | D>N | No |
ClinGen gnomAD |
|
|
rs1162965837 CA388938290 |
173 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA7106258 rs761603518 |
177 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs956500859 CA257735401 |
178 | G>R | No |
ClinGen gnomAD |
|
|
rs769518344 CA7106259 |
179 | W>R | No |
ClinGen ExAC gnomAD |
|
|
CA388938368 rs1412604370 |
181 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA388938365 rs1172779983 |
181 | R>W | No |
ClinGen TOPMed |
|
|
CA7106260 rs773741245 |
183 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA388938392 rs1179693523 |
184 | C>S | No |
ClinGen gnomAD |
|
|
CA388938411 rs1478606274 |
186 | Q>L | No |
ClinGen gnomAD |
|
|
rs763308175 CA7106261 |
188 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388938436 rs766812288 CA7106262 |
189 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA257735406 rs201408794 |
189 | D>H | No |
ClinGen Ensembl |
|
|
rs201116532 CA388938442 |
190 | A>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs201116532 CA7106263 |
190 | A>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA7106266 rs767625289 |
195 | F>C | No |
ClinGen ExAC gnomAD |
|
|
rs1335108983 CA388938487 |
195 | F>L | No |
ClinGen gnomAD |
|
|
rs1466757573 CA388938483 |
195 | F>V | No |
ClinGen gnomAD |
|
|
rs1437179282 CA388938502 |
197 | V>G | No |
ClinGen TOPMed gnomAD |
|
|
CA257735414 rs912350554 |
198 | T>I | No |
ClinGen Ensembl |
|
|
rs752752122 CA7106267 |
199 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs145460263 CA7106268 |
200 | R>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA388938522 rs145460263 |
200 | R>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs148440133 CA7106269 |
200 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA7106270 rs750231268 |
201 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7106271 rs758270170 |
201 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758270170 CA7106272 |
201 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA388938547 rs1275676703 |
203 | F>S | No |
ClinGen TOPMed |
|
|
CA388938554 rs1215696577 |
204 | S>A | No |
ClinGen TOPMed |
|
|
rs768452926 CA7106274 |
207 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA257735428 rs556260853 |
208 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs780867772 CA388938586 |
208 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs780867772 CA7106275 |
208 | E>Q | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 210 | L>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA388938623 rs1423899378 |
212 | R>Q | No |
ClinGen gnomAD |
|
|
CA7106277 rs769572112 |
214 | R>P | No |
ClinGen ExAC TOPMed |
|
|
CA388938640 rs769572112 |
214 | R>Q | No |
ClinGen ExAC TOPMed |
|
|
rs747619493 CA7106276 |
214 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA7106278 rs376686140 |
215 | A>P | No |
ClinGen ESP ExAC gnomAD |
|
|
rs376686140 CA388938645 |
215 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA gnomAD |
|
CA388938675 rs1405412559 |
218 | P>L | No |
ClinGen gnomAD |
|
|
rs748882154 CA7106279 |
218 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs774552269 CA7106281 |
219 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA388938708 rs1356092478 |
222 | L>P | No |
ClinGen TOPMed |
|
|
rs760139078 CA7106283 |
223 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs767963097 CA257735435 |
224 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7106284 rs767963097 |
224 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1595038733 CA388938732 |
225 | I>T | No |
ClinGen Ensembl |
|
|
rs1269829055 CA388938749 |
227 | V>A | No |
ClinGen gnomAD |
|
|
CA388938748 rs1269829055 |
227 | V>D | No |
ClinGen gnomAD |
|
|
rs940872931 CA257735444 |
231 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA388938803 rs1217823459 |
233 | L>F | No |
ClinGen gnomAD |
|
|
CA388938808 rs764143490 |
234 | A>D | No |
ClinGen ExAC gnomAD |
|
|
rs764143490 CA7106287 |
234 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1445632252 CA388938816 |
235 | R>H | No |
ClinGen gnomAD |
|
|
CA388938818 rs1445632252 |
235 | R>L | No |
ClinGen gnomAD |
|
|
rs1190628755 CA388938832 |
237 | R>G | No |
ClinGen gnomAD |
|
|
rs1475281221 CA388938834 |
237 | R>Q | No |
ClinGen TOPMed |
|
|
rs1472854138 CA388938850 |
239 | V>I | No |
ClinGen gnomAD |
|
|
CA257735449 rs749636718 |
241 | L>P | No |
ClinGen Ensembl |
|
|
rs1050599825 CA257735562 |
244 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1313126261 CA388938950 |
248 | A>T | No |
ClinGen TOPMed |
|
|
rs770468550 CA7106298 |
249 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1346204332 CA388938957 |
249 | G>R | No |
ClinGen gnomAD |
|
|
CA257735565 rs892408602 |
250 | T>A | No |
ClinGen Ensembl |
|
|
rs774801251 CA7106299 |
251 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA388938972 rs1460529112 |
252 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
CA388938973 rs567249320 |
252 | S>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs567249320 CA7106300 |
252 | S>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1420211441 CA388938978 |
253 | C>R | No |
ClinGen TOPMed |
|
|
rs1378056511 CA388938993 |
255 | H>N | No |
ClinGen gnomAD |
|
|
CA388939022 rs1197375345 |
259 | S>P | No |
ClinGen TOPMed |
|
|
rs1299895789 CA388939040 |
262 | L>M | No |
ClinGen gnomAD |
|
|
rs553093639 CA388939052 |
263 | H>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1394988357 CA388939049 |
263 | H>R | No |
ClinGen gnomAD |
|
|
rs952436608 CA257735579 |
266 | T>R | No |
ClinGen Ensembl |
|
| TCGA novel | 268 | E>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA257735582 rs573430135 |
271 | E>* | No |
ClinGen 1000Genomes |
|
|
rs1281007538 CA388939111 |
272 | G>C | No |
ClinGen gnomAD |
|
|
rs1281007538 CA388939110 |
272 | G>S | No |
ClinGen gnomAD |
|
|
rs1198739895 CA388939130 |
275 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA388939140 rs1252612047 |
276 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs983811743 CA257735586 |
278 | R>L | No |
ClinGen TOPMed |
|
|
CA7106307 rs762113247 |
280 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762113247 CA388939160 |
280 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1267943798 CA388939159 |
280 | R>W | No |
ClinGen gnomAD |
|
|
CA7106308 rs199599030 |
281 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1464097036 CA388939163 |
281 | R>W | No |
ClinGen TOPMed |
|
|
CA388939167 rs1163956488 |
282 | G>S | No |
ClinGen TOPMed |
|
|
CA388939172 rs1472572941 |
283 | R>G | No |
ClinGen TOPMed |
|
|
CA388939186 rs1428866825 |
285 | H>D | No |
ClinGen TOPMed gnomAD |
|
|
CA388939213 rs1413545842 |
289 | Q>* | No |
ClinGen gnomAD |
|
|
rs1371478201 CA388939221 |
290 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1461520853 CA388939228 |
291 | P>S | No |
ClinGen gnomAD |
|
|
CA388939232 rs1294684422 |
292 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
CA388939249 rs1302859974 |
294 | G>D | No |
ClinGen gnomAD |
|
|
rs1595039407 CA388939274 |
298 | G>S | No |
ClinGen Ensembl |
|
|
rs1341503776 CA388939277 |
298 | G>V | No |
ClinGen gnomAD |
|
|
CA388939287 rs1214861983 |
300 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
CA388939291 rs1297232736 |
300 | A>V | No |
ClinGen gnomAD |
|
|
CA388939297 rs1305304706 |
301 | P>L | No |
ClinGen gnomAD |
|
|
rs1225995068 CA388939302 |
302 | P>R | No |
ClinGen gnomAD |
|
|
rs559211398 CA257735593 |
304 | R>S | No |
ClinGen 1000Genomes gnomAD |
|
|
CA388939317 rs1490801103 |
305 | R>C | No |
ClinGen gnomAD |
|
|
CA388939331 rs1280942176 |
307 | S>C | No |
ClinGen TOPMed |
|
|
rs373774017 CA7106311 |
307 | S>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1303653826 CA388939334 |
307 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA388939336 rs969535392 |
308 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA257735597 rs969535392 |
308 | L>I | No |
ClinGen TOPMed gnomAD |
|
|
CA388939346 rs1417613685 |
309 | T>I | No |
ClinGen gnomAD |
|
| TCGA novel | 312 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA257735599 rs772787398 |
314 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1373876149 CA388939397 |
317 | A>T | No |
ClinGen TOPMed |
|
|
rs1324276312 CA388939407 |
318 | N>S | No |
ClinGen TOPMed |
|
|
CA388939424 rs1163498459 |
321 | P>S | No |
ClinGen gnomAD |
|
|
rs1433295616 CA388939457 |
326 | F>V | No |
ClinGen gnomAD |
|
|
rs767409341 CA7106312 |
329 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA388939492 rs1409273943 |
330 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
CA388939503 rs1286471654 |
332 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
rs368798364 CA257735602 |
332 | R>T | No |
ClinGen ESP |
|
|
CA7106313 rs752750738 |
338 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1290909293 CA388939549 |
339 | V>E | No |
ClinGen gnomAD |
|
|
rs972702237 CA257735606 |
341 | L>C | No |
ClinGen Ensembl |
No associated diseases with Q8IYK8
1 regional properties for Q8IYK8
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| binding_site | Sigma-54 interaction domain, ATP-binding site 1 | 117 - 130 | IPR025662 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium channel regulator activity | Modulates the activity of a calcium channel. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
34 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P08642 | HRAS | GTPase HRas | Gallus gallus (Chicken) | SS |
| P11233 | RALA | Ras-related protein Ral-A | Homo sapiens (Human) | PR |
| Q6T310 | RASL11A | Ras-like protein family member 11A | Homo sapiens (Human) | PR |
| P01116 | KRAS | GTPase KRas | Homo sapiens (Human) | EV |
| P62070 | RRAS2 | Ras-related protein R-Ras2 | Homo sapiens (Human) | PR |
| P11234 | RALB | Ras-related protein Ral-B | Homo sapiens (Human) | PR |
| Q99578 | RIT2 | GTP-binding protein Rit2 | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q9BU20 | CPLANE2 | Ciliogenesis and planar polarity effector 2 | Homo sapiens (Human) | PR |
| Q96HU8 | DIRAS2 | GTP-binding protein Di-Ras2 | Homo sapiens (Human) | PR |
| P01112 | HRAS | GTPase HRas | Homo sapiens (Human) | SS |
| P55040 | GEM | GTP-binding protein GEM | Homo sapiens (Human) | PR |
| Q9JIW9 | Ralb | Ras-related protein Ral-B | Mus musculus (Mouse) | PR |
| Q61411 | Hras | GTPase HRas | Mus musculus (Mouse) | SS |
| P32883 | Kras | GTPase KRas | Mus musculus (Mouse) | SS |
| O08989 | Mras | Ras-related protein M-Ras | Mus musculus (Mouse) | PR |
| Q5PR73 | Diras2 | GTP-binding protein Di-Ras2 | Mus musculus (Mouse) | PR |
| Q91Z61 | Diras1 | GTP-binding protein Di-Ras1 | Mus musculus (Mouse) | PR |
| P62071 | Rras2 | Ras-related protein R-Ras2 | Mus musculus (Mouse) | PR |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| Q08AT1 | Rasl12 | Ras-like protein family member 12 | Mus musculus (Mouse) | PR |
| A2A825 | Cplane2 | Ciliogenesis and planar polarity effector 2 | Mus musculus (Mouse) | PR |
| P70425 | Rit2 | GTP-binding protein Rit2 | Mus musculus (Mouse) | PR |
| P55041 | Gem | GTP-binding protein GEM | Mus musculus (Mouse) | PR |
| Q8VEL9 | Rem2 | GTP-binding protein REM 2 | Mus musculus (Mouse) | PR |
| P36860 | Ralb | Ras-related protein Ral-B | Rattus norvegicus (Rat) | PR |
| P08644 | Kras | GTPase KRas | Rattus norvegicus (Rat) | SS |
| P20171 | Hras | GTPase HRas | Rattus norvegicus (Rat) | SS |
| Q5BJQ5 | Rit2 | GTP-binding protein Rit2 | Rattus norvegicus (Rat) | PR |
| P97538 | Mras | Ras-related protein M-Ras | Rattus norvegicus (Rat) | PR |
| Q9WTY2 | Rem2 | GTP-binding protein REM 2 | Rattus norvegicus (Rat) | PR |
| B7ZTR0 | cplane2 | Ciliogenesis and planar polarity effector 2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| P79737 | nras | GTPase NRas | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A1DZY4 | zgc:110179 | Ras-like protein family member 11A-like | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MHTDLDTDMD | MDTETTALCP | SGSRRASPPG | TPTPEADATL | LKKSEKLLAE | LDRSGLPSAP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GAPRRRGSMP | VPYKHQLRRA | QAVDELDWPP | QASSSGSSDS | LGSGEAAPAQ | KDGIFKVMLV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GESGVGKSTL | AGTFGGLQGD | SAHEPENPED | TYERRIMVDK | EEVTLVVYDI | WEQGDAGGWL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RDHCLQTGDA | FLIVFSVTDR | RSFSKVPETL | LRLRAGRPHH | DLPVILVGNK | SDLARSREVS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LEEGRHLAGT | LSCKHIETSA | ALHHNTRELF | EGAVRQIRLR | RGRNHAGGQR | PDPGSPEGPA |
| 310 | 320 | 330 | |||
| PPARRESLTK | KAKRFLANLV | PRNAKFFKQR | SRSCHDLSVL |