Q8IXJ6
Gene name |
SIRT2 (SIR2L, SIR2L2) |
Protein name |
NAD-dependent protein deacetylase sirtuin-2 |
Names |
Regulatory protein SIR2 homolog 2, SIR2-like protein 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:22933 |
EC number |
2.3.1.286: Transferring groups other than amino-acyl groups |
Protein Class |
NAD-DEPENDENT PROTEIN DEACYLASE SIRTUIN-5, MITOCHONDRIAL-RELATED (PTHR11085) |
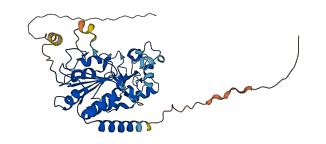
Descriptions
Sirtuin 2 (SIRT2) is a NAD+ dependent deacetylase that has been associated with neurodegeneration and cancer. The C-terminal region (CT) of SIRT2 functions as an autoinhibitory region that regulates the deacetylation activity of SIRT2. Phosphorylation at S331 of SIRT2 isoform 2 causes large conformational changes in the CT that enhance the autoinhibitory activity of the CT region. This serine residue is located within the naturally disordered C-terminal region (CT, residues 320-352 in isoform 2).
Autoinhibitory domains (AIDs)
Target domain |
26-319 (Catalytic core domain) |
Relief mechanism |
Others |
Assay |
Deletion assay, Structural analysis |
Accessory elements
No accessory elements
References
- Li J et al. (2015) "Insight into the Mechanism of Intramolecular Inhibition of the Catalytic Activity of Sirtuin 2 (SIRT2)", PloS one, 10, e0139095
- Xu Y et al. (2014) "Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase", Cancer research, 74, 3630-42
- Finnin MS et al. (2001) "Structure of the histone deacetylase SIRT2", Nature structural biology, 8, 621-5
Autoinhibited structure
Activated structure

40 structures for Q8IXJ6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1J8F | X-ray | 170 A | A/B/C | 34-356 | PDB |
| 3ZGO | X-ray | 163 A | A/B/C | 34-356 | PDB |
| 3ZGV | X-ray | 227 A | A/B | 34-356 | PDB |
| 4L3O | X-ray | 252 A | A/B/C/D | 55-356 | PDB |
| 4R8M | X-ray | 210 A | A/B | 38-356 | PDB |
| 4RMG | X-ray | 188 A | A | 56-356 | PDB |
| 4RMH | X-ray | 142 A | A | 56-356 | PDB |
| 4RMI | X-ray | 145 A | A | 56-356 | PDB |
| 4RMJ | X-ray | 187 A | A/B | 56-356 | PDB |
| 4X3O | X-ray | 150 A | A | 52-355 | PDB |
| 4X3P | X-ray | 180 A | A | 52-355 | PDB |
| 4Y6L | X-ray | 160 A | A/B | 52-356 | PDB |
| 4Y6O | X-ray | 160 A | A/B | 52-356 | PDB |
| 4Y6Q | X-ray | 190 A | A/B/C/D | 52-356 | PDB |
| 5D7O | X-ray | 163 A | A/B | 50-356 | PDB |
| 5D7P | X-ray | 176 A | A/B | 56-356 | PDB |
| 5D7Q | X-ray | 201 A | A/B | 56-356 | PDB |
| 5DY4 | X-ray | 177 A | A | 56-356 | PDB |
| 5DY5 | X-ray | 195 A | A | 56-356 | PDB |
| 5FYQ | X-ray | 300 A | A/B | 1-356 | PDB |
| 5G4C | X-ray | 210 A | A/B | 34-356 | PDB |
| 5MAR | X-ray | 189 A | A/B | 56-356 | PDB |
| 5MAT | X-ray | 207 A | A/C | 56-356 | PDB |
| 5Y0Z | X-ray | 200 A | A/B | 52-356 | PDB |
| 5Y5N | X-ray | 230 A | A | 32-356 | PDB |
| 5YQL | X-ray | 160 A | A | 56-356 | PDB |
| 5YQM | X-ray | 174 A | A | 56-356 | PDB |
| 5YQN | X-ray | 160 A | A | 56-356 | PDB |
| 5YQO | X-ray | 148 A | A | 56-356 | PDB |
| 6L65 | X-ray | 180 A | A | 50-355 | PDB |
| 6L66 | X-ray | 217 A | A | 52-355 | PDB |
| 6L71 | X-ray | 211 A | A | 52-355 | PDB |
| 6L72 | X-ray | 250 A | A | 52-355 | PDB |
| 6NR0 | X-ray | 245 A | A/B | 38-356 | PDB |
| 6QCN | X-ray | 223 A | A/B | 55-356 | PDB |
| 7BOS | X-ray | 170 A | A | 52-356 | PDB |
| 7BOT | X-ray | 170 A | A | 52-356 | PDB |
| 8OWZ | X-ray | 165 A | A | 56-356 | PDB |
| 8TGP | X-ray | 176 A | A | 34-356 | PDB |
| AF-Q8IXJ6-F1 | Predicted | AlphaFoldDB |
688 variants for Q8IXJ6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1431209574 | 5 | D>N | No | gnomAD | |
|
CA405730788 rs1431209574 |
5 | D>N | No |
ClinGen gnomAD |
|
| rs748872953 | 6 | P>A | No |
ExAC TOPMed gnomAD |
|
|
CA9422092 rs748872953 |
6 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs904071840 | 6 | P>L | No |
TOPMed gnomAD |
|
|
CA308161239 rs904071840 |
6 | P>L | No |
ClinGen TOPMed gnomAD |
|
| rs774995182 | 8 | H>Y | No |
ExAC gnomAD |
|
|
rs774995182 CA9422072 |
8 | H>Y | No |
ClinGen ExAC gnomAD |
|
| rs1362615136 | 9 | P>T | No | gnomAD | |
|
CA405730753 rs1362615136 |
9 | P>T | No |
ClinGen gnomAD |
|
| rs1418810049 | 12 | T>I | No |
TOPMed gnomAD |
|
|
CA405730729 rs1418810049 |
12 | T>I | No |
ClinGen TOPMed gnomAD |
|
| rs1473123135 | 14 | A>P | No |
TOPMed gnomAD |
|
|
CA405730718 rs1473123135 |
14 | A>P | No |
ClinGen TOPMed gnomAD |
|
| rs776314748 | 17 | V>E | No |
ExAC gnomAD |
|
|
CA9422069 rs776314748 |
17 | V>E | No |
ClinGen ExAC gnomAD |
|
| rs1428015087 | 17 | V>M | No | gnomAD | |
|
CA405730700 rs1428015087 |
17 | V>M | No |
ClinGen gnomAD |
|
|
rs201682459 CA9422068 |
18 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs201682459 | 18 | Q>H | No |
ExAC TOPMed gnomAD |
|
|
rs1293047926 CA405730692 |
18 | Q>P | No |
ClinGen TOPMed |
|
| rs1293047926 | 18 | Q>P | No | TOPMed | |
|
rs1600134981 CA405730680 |
20 | A>T | No |
ClinGen Ensembl |
|
| rs1600134981 | 20 | A>T | No | Ensembl | |
| rs777560427 | 20 | A>V | No |
ExAC gnomAD |
|
|
CA9422066 rs777560427 |
20 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs370835884 CA9422019 |
22 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs370835884 | 22 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
|
rs370835884 CA308157797 |
22 | D>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs370835884 | 22 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs758838464 | 23 | S>L | No |
ExAC gnomAD |
|
|
rs758838464 CA9422017 |
23 | S>L | No |
ClinGen ExAC gnomAD |
|
| rs765827093 | 25 | S>* | No |
ExAC gnomAD |
|
|
CA9422015 rs765827093 |
25 | S>* | No |
ClinGen ExAC gnomAD |
|
|
rs765827093 CA405730582 |
25 | S>L | No |
ClinGen ExAC gnomAD |
|
| rs765827093 | 25 | S>L | No |
ExAC gnomAD |
|
| rs996409143 | 26 | D>G | No | gnomAD | |
|
rs996409143 CA308157791 |
26 | D>G | No |
ClinGen gnomAD |
|
| rs200832504 | 27 | S>C | No |
ESP ExAC TOPMed gnomAD |
|
|
rs200832504 CA9422014 |
27 | S>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs1600127235 | 28 | E>G | No | Ensembl | |
|
rs1600127235 CA405730565 |
28 | E>G | No |
ClinGen Ensembl |
|
|
CA308157782 rs748102696 |
30 | G>E | No |
ClinGen TOPMed |
|
| rs748102696 | 30 | G>E | No | TOPMed | |
|
rs748102696 CA405730551 |
30 | G>V | No |
ClinGen TOPMed |
|
| rs748102696 | 30 | G>V | No | TOPMed | |
| rs1019096585 | 31 | A>S | No | Ensembl | |
|
CA308157773 rs1019096585 |
31 | A>S | No |
ClinGen Ensembl |
|
| rs1019096585 | 31 | A>T | No | Ensembl | |
|
CA308157776 rs1019096585 |
31 | A>T | No |
ClinGen Ensembl |
|
| rs926292218 | 32 | A>T | No |
TOPMed gnomAD |
|
|
COSM996185 rs926292218 CA308157764 |
32 | A>T | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
| rs767144427 | 32 | A>V | No |
ExAC gnomAD |
|
|
rs767144427 CA9422010 |
32 | A>V | No |
ClinGen ExAC gnomAD |
|
| rs199515794 | 34 | G>R | No |
ExAC TOPMed gnomAD |
|
|
CA9422009 rs199515794 |
34 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs1460165551 | 36 | A>E | No | gnomAD | |
|
CA405730520 rs1460165551 |
36 | A>E | No |
ClinGen gnomAD |
|
|
CA405730494 rs1172707908 |
38 | M>T | No |
ClinGen gnomAD |
|
| rs1172707908 | 38 | M>T | No | gnomAD | |
|
CA9422008 rs201344394 |
38 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs201344394 | 38 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs200012062 | 42 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs200012062 CA9421967 |
42 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs765083798 | 42 | R>W | No |
ExAC TOPMed gnomAD |
|
|
rs765083798 CA9421968 |
42 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9421966 RCV000955223 rs45535036 |
44 | L>F | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs45535036 | 44 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200819077 | 45 | F>L | No | Ensembl | |
|
rs200819077 CA308157326 |
45 | F>L | No |
ClinGen Ensembl |
|
|
rs376694959 CA9421965 |
48 | T>M | No |
ClinGen ESP ExAC gnomAD |
|
| rs376694959 | 48 | T>M | No |
ESP ExAC gnomAD |
|
| rs1201810649 | 49 | L>I | No | gnomAD | |
|
CA405730424 rs1201810649 |
49 | L>I | No |
ClinGen gnomAD |
|
|
CA9421963 rs546115964 |
50 | S>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| rs546115964 | 50 | S>G | No |
1000Genomes ExAC gnomAD |
|
|
CA308157303 rs543937371 |
51 | L>P | No |
ClinGen Ensembl |
|
| rs543937371 | 51 | L>P | No | Ensembl | |
| rs575999768 | 51 | L>V | No |
1000Genomes ExAC gnomAD |
|
|
rs575999768 CA9421962 |
51 | L>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA9421961 rs762039728 |
55 | K>N | No |
ClinGen ExAC gnomAD |
|
| rs762039728 | 55 | K>N | No |
ExAC gnomAD |
|
|
CA9421959 rs139357839 |
57 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs139357839 | 57 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs139357839 | 57 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs139357839 CA405730369 |
57 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs774515802 | 57 | R>S | No |
ExAC TOPMed gnomAD |
|
|
CA9421960 rs774515802 |
57 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs749731782 | 60 | D>N | No |
ExAC TOPMed gnomAD |
|
|
rs749731782 CA9421958 |
60 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs200888217 | 61 | E>D | No | Ensembl | |
|
CA308157270 rs200888217 |
61 | E>D | No |
ClinGen Ensembl |
|
| rs920534440 | 61 | E>K | No | Ensembl | |
|
rs920534440 CA308157277 |
61 | E>K | No |
ClinGen Ensembl |
|
| rs1403129868 | 62 | L>P | No | gnomAD | |
|
rs1403129868 CA405730342 |
62 | L>P | No |
ClinGen gnomAD |
|
| rs1034418160 | 64 | L>F | No |
TOPMed gnomAD |
|
|
rs1034418160 CA308157255 |
64 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA405730328 rs1600126259 |
65 | E>K | No |
ClinGen Ensembl |
|
| rs1600126259 | 65 | E>K | No | Ensembl | |
| rs770557472 | 66 | G>V | No |
ExAC gnomAD |
|
|
CA9421956 rs770557472 |
66 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA405730315 rs1600126238 |
67 | V>M | No |
ClinGen Ensembl |
|
| rs1600126238 | 67 | V>M | No | Ensembl | |
| rs201911244 | 69 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs201911244 CA405730301 |
69 | R>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs201911244 | 69 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs201911244 CA9421953 |
69 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs199676461 | 69 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421954 rs199676461 |
69 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs764664021 | 70 | Y>D | No | Ensembl | |
|
CA308157229 rs764664021 |
70 | Y>D | No |
ClinGen Ensembl |
|
| rs1037998278 | 71 | M>I | No | TOPMed | |
|
rs1037998278 CA308157223 |
71 | M>I | No |
ClinGen TOPMed |
|
|
CA9421952 rs752424589 |
71 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs752424589 | 71 | M>L | No |
ExAC TOPMed gnomAD |
|
|
rs752424589 CA405730292 |
71 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs752424589 | 71 | M>V | No |
ExAC TOPMed gnomAD |
|
|
CA308157210 rs376265776 |
73 | S>N | No |
ClinGen ESP TOPMed gnomAD |
|
| rs376265776 | 73 | S>N | No |
ESP TOPMed gnomAD |
|
|
rs200563255 CA9421951 COSM711535 |
73 | S>R | lung [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
| rs200563255 | 73 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421949 rs199594239 |
74 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs199594239 | 74 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421948 rs201405136 |
75 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs201405136 | 75 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs200520235 | 75 | R>H | No |
ExAC TOPMed gnomAD |
|
|
rs200520235 CA9421947 |
75 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405730244 rs1351485075 |
76 | C>F | No |
ClinGen TOPMed |
|
| rs1351485075 | 76 | C>F | No | TOPMed | |
|
rs146760070 CA9421911 |
77 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs146760070 | 77 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs146760070 | 77 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
|
rs146760070 CA308155241 |
77 | R>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs773864674 | 77 | R>H | No |
ExAC TOPMed gnomAD |
|
|
rs773864674 CA9421910 |
77 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs773864674 CA9421909 |
77 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs773864674 | 77 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs773864674 | 77 | R>P | No |
ExAC TOPMed gnomAD |
|
|
rs773864674 CA308155230 |
77 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1360499204 CA405730234 |
78 | R>S | No |
ClinGen TOPMed gnomAD |
|
| rs1360499204 | 78 | R>S | No |
TOPMed gnomAD |
|
|
rs1600120923 CA405730227 |
80 | I>F | No |
ClinGen Ensembl |
|
| rs1600120923 | 80 | I>F | No | Ensembl | |
| rs1455261976 | 80 | I>M | No | TOPMed | |
|
CA405730221 rs1455261976 |
80 | I>M | No |
ClinGen TOPMed |
|
|
CA308155219 rs373609953 |
83 | V>L | No |
ClinGen ESP |
|
| rs373609953 | 83 | V>L | No | ESP | |
| rs143528673 | 87 | I>F | No |
ESP ExAC TOPMed gnomAD |
|
|
rs143528673 CA9421906 |
87 | I>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs143528673 | 87 | I>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs143528673 CA405730182 |
87 | I>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs1453825601 | 88 | S>A | No | TOPMed | |
|
rs1453825601 CA405730174 |
88 | S>A | No |
ClinGen TOPMed |
|
| rs934433059 | 88 | S>F | No | TOPMed | |
|
CA308155206 rs934433059 |
88 | S>F | No |
ClinGen TOPMed |
|
| rs750920243 | 90 | S>F | No |
ExAC TOPMed gnomAD |
|
|
rs750920243 CA9421875 |
90 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201249832 CA308155204 |
90 | S>P | No |
ClinGen gnomAD |
|
| rs201249832 | 90 | S>P | No | gnomAD | |
|
CA9421872 rs200814071 |
91 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs200814071 | 91 | A>S | No |
ExAC TOPMed gnomAD |
|
|
CA9421873 rs200814071 |
91 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs200814071 | 91 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1428868182 | 95 | D>H | No |
TOPMed gnomAD |
|
|
rs1428868182 CA405730125 |
95 | D>H | No |
ClinGen TOPMed gnomAD |
|
|
CA405730126 rs1428868182 |
95 | D>N | No |
ClinGen TOPMed gnomAD |
|
| rs1428868182 | 95 | D>N | No |
TOPMed gnomAD |
|
|
rs1469719084 CA405730111 |
96 | F>L | No |
ClinGen TOPMed gnomAD |
|
| rs1469719084 | 96 | F>L | No |
TOPMed gnomAD |
|
|
rs1184695675 CA405730116 |
96 | F>V | No |
ClinGen gnomAD |
|
| rs1184695675 | 96 | F>V | No | gnomAD | |
| rs769376141 | 97 | R>C | No |
ExAC TOPMed gnomAD |
|
|
CA308155099 rs769376141 |
97 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs769376141 | 97 | R>G | No |
ExAC TOPMed gnomAD |
|
|
CA405730110 rs769376141 |
97 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs374491281 | 97 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421869 rs374491281 |
97 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9421870 rs769376141 |
97 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs769376141 | 97 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs1227795348 | 98 | S>C | No | gnomAD | |
|
CA405730103 rs1227795348 |
98 | S>C | No |
ClinGen gnomAD |
|
|
CA405730106 rs1259551409 |
98 | S>P | No |
ClinGen gnomAD |
|
| rs1259551409 | 98 | S>P | No | gnomAD | |
| rs1295226901 | 99 | P>L | No | gnomAD | |
|
rs1295226901 CA405730097 |
99 | P>L | No |
ClinGen gnomAD |
|
| rs776368661 | 99 | P>S | No |
ExAC gnomAD |
|
|
CA9421868 rs776368661 |
99 | P>S | No |
ClinGen ExAC gnomAD |
|
| rs770654234 | 100 | S>Y | No |
ExAC gnomAD |
|
|
CA9421867 rs770654234 |
100 | S>Y | No |
ClinGen ExAC gnomAD |
|
| rs1482531642 | 102 | G>D | No | TOPMed | |
|
rs1482531642 CA405730084 |
102 | G>D | No |
ClinGen TOPMed |
|
|
CA9421865 rs76265208 |
102 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs76265208 | 102 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs76265208 | 102 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
CA9421864 rs76265208 |
102 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA405730079 rs1440239604 |
103 | L>F | No |
ClinGen gnomAD |
|
| rs1440239604 | 103 | L>F | No | gnomAD | |
|
CA9421863 rs748298105 |
104 | Y>C | No |
ClinGen ExAC gnomAD |
|
| rs748298105 | 104 | Y>C | No |
ExAC gnomAD |
|
| rs1300115367 | 106 | N>D | No | gnomAD | |
|
CA405730059 rs1300115367 |
106 | N>D | No |
ClinGen gnomAD |
|
|
rs1462742971 CA405730056 |
106 | N>S | No |
ClinGen gnomAD |
|
| rs1462742971 | 106 | N>S | No | gnomAD | |
|
CA9421862 rs779149028 |
107 | L>I | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs779149028 | 107 | L>I | No |
ExAC TOPMed gnomAD |
|
|
rs551071804 CA9421861 |
108 | E>K | No |
ClinGen ExAC gnomAD |
|
| rs551071804 | 108 | E>K | No |
ExAC gnomAD |
|
| rs200159438 | 111 | H>R | No |
ExAC gnomAD |
|
|
rs200159438 CA9421860 |
111 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs780421129 CA9421859 |
113 | P>L | No |
ClinGen ExAC gnomAD |
|
| rs780421129 | 113 | P>L | No |
ExAC gnomAD |
|
|
rs1466746052 CA405730011 |
113 | P>T | No |
ClinGen gnomAD |
|
| rs1466746052 | 113 | P>T | No | gnomAD | |
| rs200307334 | 115 | P>L | No |
ExAC TOPMed gnomAD |
|
|
rs200307334 CA9421858 |
115 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA308155040 rs11551762 |
117 | A>S | No |
ClinGen Ensembl |
|
| rs11551762 | 117 | A>S | No | Ensembl | |
| rs199610453 | 120 | E>D | No |
ExAC TOPMed gnomAD |
|
|
CA9421856 rs199610453 |
120 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs1389738435 | 120 | E>K | No | TOPMed | |
|
rs1389738435 CA405729966 |
120 | E>K | No |
ClinGen TOPMed |
|
|
CA308155024 rs201156500 |
122 | S>N | No |
ClinGen gnomAD |
|
| rs201156500 | 122 | S>N | No | gnomAD | |
| rs200366538 | 124 | F>L | No |
ExAC TOPMed gnomAD |
|
|
CA9421854 rs200366538 |
124 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs199856206 | 128 | P>L | No |
ExAC TOPMed gnomAD |
|
|
COSM996184 rs199856206 CA9421826 |
128 | P>L | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
| rs1441267044 | 129 | E>A | No | TOPMed | |
|
rs1441267044 CA405729887 |
129 | E>A | No |
ClinGen TOPMed |
|
| rs1206306591 | 130 | P>S | No | gnomAD | |
|
CA405729880 rs1206306591 |
130 | P>S | No |
ClinGen gnomAD |
|
|
rs775649914 CA308154888 |
131 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs775649914 | 131 | F>L | No |
ExAC TOPMed gnomAD |
|
|
CA9421824 rs749382036 |
131 | F>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs749382036 | 131 | F>V | No |
ExAC TOPMed gnomAD |
|
|
rs1600119545 CA405729868 |
132 | F>V | No |
ClinGen Ensembl |
|
| rs1600119545 | 132 | F>V | No | Ensembl | |
|
CA9421820 rs150309258 |
133 | A>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150309258 | 133 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs150309258 CA9421821 |
133 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150309258 | 133 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs757666044 | 133 | A>V | No |
ExAC gnomAD |
|
|
rs757666044 CA9421819 |
133 | A>V | No |
ClinGen ExAC gnomAD |
|
| rs143889239 | 135 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs143889239 CA9421817 |
135 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA9421816 rs374519216 |
136 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs374519216 | 136 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421815 rs753231190 |
137 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs753231190 | 137 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs755684272 | 140 | P>L | No |
ExAC TOPMed gnomAD |
|
|
CA9421813 rs755684272 |
140 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs1029032321 | 143 | F>L | No | Ensembl | |
|
CA308154843 rs1029032321 |
143 | F>L | No |
ClinGen Ensembl |
|
| rs1477759874 | 143 | F>L | No | TOPMed | |
|
rs1477759874 CA405729800 |
143 | F>L | No |
ClinGen TOPMed |
|
|
rs1177790257 CA405729790 |
144 | K>R | No |
ClinGen TOPMed |
|
| rs1177790257 | 144 | K>R | No | TOPMed | |
| rs906959018 | 146 | T>I | No | TOPMed | |
|
CA308154594 rs906959018 |
146 | T>I | No |
ClinGen TOPMed |
|
|
rs772516388 CA9421782 |
147 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs772516388 | 147 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1250193381 | 148 | C>S | No | TOPMed | |
|
CA405729758 rs1250193381 |
148 | C>S | No |
ClinGen TOPMed |
|
| rs779520276 | 149 | H>R | No |
ExAC TOPMed gnomAD |
|
|
CA9421780 rs779520276 |
149 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs1472469269 | 149 | H>Y | No | TOPMed | |
|
rs1472469269 CA405729748 |
149 | H>Y | No |
ClinGen TOPMed |
|
|
CA405729740 rs1313246046 |
150 | Y>C | No |
ClinGen gnomAD |
|
| rs1313246046 | 150 | Y>C | No | gnomAD | |
| rs868272193 | 151 | F>L | No | gnomAD | |
|
CA308154589 rs868272193 |
151 | F>L | No |
ClinGen gnomAD |
|
| rs1158692353 | 152 | M>T | No | gnomAD | |
|
CA405729724 rs1158692353 |
152 | M>T | No |
ClinGen gnomAD |
|
| rs1471694390 | 153 | R>C | No | gnomAD | |
|
COSM996182 rs1471694390 CA405729719 |
153 | R>C | endometrium [Cosmic] | No |
ClinGen cosmic curated gnomAD |
| rs144373891 | 153 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV000966482 CA9421778 rs144373891 |
153 | R>H | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs780865991 | 157 | D>A | No |
ExAC gnomAD |
|
|
CA9421777 rs780865991 |
157 | D>A | No |
ClinGen ExAC gnomAD |
|
|
rs1485248024 CA405729694 |
157 | D>Y | No |
ClinGen gnomAD |
|
| rs1485248024 | 157 | D>Y | No | gnomAD | |
|
CA9421776 rs756924846 |
158 | K>Q | No |
ClinGen ExAC gnomAD |
|
| rs756924846 | 158 | K>Q | No |
ExAC gnomAD |
|
| rs1316970707 | 159 | G>R | No | gnomAD | |
|
rs1316970707 CA405729680 |
159 | G>R | No |
ClinGen gnomAD |
|
| rs1568401521 | 161 | L>F | No | Ensembl | |
|
CA405729669 rs1568401521 |
161 | L>F | No |
ClinGen Ensembl |
|
|
rs1049750023 CA308154582 |
163 | R>C | No |
ClinGen TOPMed gnomAD |
|
| rs1049750023 | 163 | R>C | No |
TOPMed gnomAD |
|
|
rs1049750023 CA405729659 |
163 | R>G | No |
ClinGen TOPMed gnomAD |
|
| rs1049750023 | 163 | R>G | No |
TOPMed gnomAD |
|
| rs751291175 | 163 | R>H | No |
ExAC TOPMed gnomAD |
|
|
CA9421775 rs751291175 |
163 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs777682267 | 165 | Y>N | No |
ExAC gnomAD |
|
|
CA9421774 rs777682267 |
165 | Y>N | No |
ClinGen ExAC gnomAD |
|
| rs752565046 | 166 | T>M | No |
ExAC TOPMed gnomAD |
|
|
rs752565046 CA9421772 |
166 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs1158466489 | 167 | Q>K | No | gnomAD | |
|
rs1158466489 CA405729636 |
167 | Q>K | No |
ClinGen gnomAD |
|
| rs1195377700 | 169 | I>L | No | TOPMed | |
|
rs1195377700 CA405729606 |
169 | I>L | No |
ClinGen TOPMed |
|
| rs975774968 | 169 | I>T | No | TOPMed | |
|
rs975774968 CA308151791 |
169 | I>T | No |
ClinGen TOPMed |
|
|
rs200906111 CA9421753 |
170 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs200906111 | 170 | D>G | No |
ExAC TOPMed gnomAD |
|
|
CA308151790 rs550825769 |
171 | T>N | No |
ClinGen 1000Genomes |
|
| rs550825769 | 171 | T>N | No | 1000Genomes | |
| rs200871138 | 174 | R>* | No |
1000Genomes ExAC gnomAD |
|
|
rs200871138 CA9421751 |
174 | R>* | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA9421750 rs756233396 |
174 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs756233396 | 174 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1226914494 | 176 | A>V | No | gnomAD | |
|
CA405729563 rs1226914494 |
176 | A>V | No |
ClinGen gnomAD |
|
| rs1398581419 | 177 | G>A | No | TOPMed | |
|
CA405729558 rs1398581419 |
177 | G>A | No |
ClinGen TOPMed |
|
|
CA405729561 rs74490433 |
177 | G>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs74490433 | 177 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs74490433 | 177 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs74490433 CA9421749 |
177 | G>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1388786657 | 180 | Q>* | No | gnomAD | |
|
CA405729543 rs1388786657 |
180 | Q>* | No |
ClinGen gnomAD |
|
| rs761942195 | 180 | Q>R | No |
ExAC gnomAD |
|
|
rs761942195 CA9421747 |
180 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1394523935 CA405729523 |
182 | D>E | No |
ClinGen TOPMed |
|
| rs1394523935 | 182 | D>E | No | TOPMed | |
|
rs764400439 CA9421745 |
182 | D>N | No |
ClinGen ExAC gnomAD |
|
| rs764400439 | 182 | D>N | No |
ExAC gnomAD |
|
| rs1166630978 | 184 | V>M | No | gnomAD | |
|
CA405729514 rs1166630978 |
184 | V>M | No |
ClinGen gnomAD |
|
|
rs201307475 CA9421744 |
185 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs201307475 | 185 | E>Q | No |
ExAC TOPMed gnomAD |
|
|
CA405729496 rs1324531579 COSM996181 |
186 | A>V | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
| rs1324531579 | 186 | A>V | No | TOPMed | |
|
rs1430999088 CA405729487 |
188 | G>S | No |
ClinGen gnomAD |
|
| rs1430999088 | 188 | G>S | No | gnomAD | |
|
rs1600109632 CA405729481 |
189 | T>P | No |
ClinGen Ensembl |
|
| rs1600109632 | 189 | T>P | No | Ensembl | |
| rs771697414 | 194 | H>Q | No |
ExAC gnomAD |
|
|
rs771697414 CA9421739 |
194 | H>Q | No |
ClinGen ExAC gnomAD |
|
| rs377197576 | 194 | H>Y | No | Ensembl | |
|
rs377197576 CA308151788 |
194 | H>Y | No |
ClinGen Ensembl |
|
| rs1203459352 | 195 | C>F | No | gnomAD | |
|
CA405729438 rs1203459352 |
195 | C>F | No |
ClinGen gnomAD |
|
|
rs202089013 CA9421737 |
195 | C>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| rs202089013 | 195 | C>G | No |
1000Genomes ExAC gnomAD |
|
| rs202089013 | 195 | C>R | No |
1000Genomes ExAC gnomAD |
|
|
CA9421738 rs202089013 |
195 | C>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| rs200616294 | 196 | V>I | No |
ESP TOPMed gnomAD |
|
|
CA308151787 COSM1393477 rs200616294 |
196 | V>I | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP TOPMed gnomAD |
| rs200503228 | 198 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421733 rs200503228 |
198 | A>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
RCV000962683 CA9421734 rs34321258 |
198 | A>T | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs34321258 | 198 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200503228 | 198 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
|
rs200503228 CA9421732 |
198 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs767520526 | 200 | C>Y | No |
ExAC TOPMed |
|
|
CA9421731 rs767520526 |
200 | C>Y | No |
ClinGen ExAC TOPMed |
|
| rs756288624 | 201 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs756288624 CA9421729 |
201 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9421730 rs199646012 |
201 | R>W | No |
ClinGen ExAC gnomAD |
|
| rs199646012 | 201 | R>W | No |
ExAC gnomAD |
|
|
CA9421727 rs200953200 |
203 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs200953200 | 203 | E>K | No |
ExAC TOPMed gnomAD |
|
|
rs1418097146 CA405729377 |
205 | P>A | No |
ClinGen TOPMed |
|
| rs1418097146 | 205 | P>A | No | TOPMed | |
|
rs776137646 CA9421726 |
205 | P>L | No |
ClinGen ExAC gnomAD |
|
| rs776137646 | 205 | P>L | No |
ExAC gnomAD |
|
| rs1418097146 | 205 | P>T | No | TOPMed | |
|
CA405729378 rs1418097146 |
205 | P>T | No |
ClinGen TOPMed |
|
| rs1171962029 | 207 | S>R | No |
TOPMed gnomAD |
|
|
CA405729361 rs1171962029 |
207 | S>R | No |
ClinGen TOPMed gnomAD |
|
| rs765774257 | 208 | W>R | No |
ExAC gnomAD |
|
|
CA9421725 rs765774257 |
208 | W>R | No |
ClinGen ExAC gnomAD |
|
|
rs146903375 CA9421724 |
209 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs146903375 | 209 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
|
rs372384472 CA405729338 |
210 | K>N | No |
ClinGen gnomAD |
|
| rs372384472 | 210 | K>N | No | gnomAD | |
|
CA9421723 rs772729178 |
211 | E>* | No |
ClinGen ExAC gnomAD |
|
| rs772729178 | 211 | E>* | No |
ExAC gnomAD |
|
|
rs570453680 CA405729311 |
212 | K>N | No |
ClinGen 1000Genomes gnomAD |
|
|
rs570453680 CA405729312 |
212 | K>N | No |
ClinGen 1000Genomes gnomAD |
|
| rs570453680 | 212 | K>N | No |
1000Genomes gnomAD |
|
| rs570453680 | 212 | K>N | No |
1000Genomes gnomAD |
|
| rs774045867 | 213 | I>F | No |
ExAC gnomAD |
|
|
CA405729309 rs774045867 |
213 | I>F | No |
ClinGen ExAC gnomAD |
|
| rs774045867 | 213 | I>V | No |
ExAC gnomAD |
|
|
rs774045867 CA9421703 |
213 | I>V | No |
ClinGen ExAC gnomAD |
|
| rs1450081520 | 215 | S>Y | No | TOPMed | |
|
CA405729293 rs1450081520 |
215 | S>Y | No |
ClinGen TOPMed |
|
|
CA9421701 rs116099388 |
217 | V>M | No |
ClinGen 1000Genomes ExAC TOPMed |
|
| rs116099388 | 217 | V>M | No |
1000Genomes ExAC TOPMed |
|
| rs762603207 | 218 | T>A | No |
ExAC TOPMed gnomAD |
|
|
CA9421700 rs762603207 |
218 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs142022596 | 218 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs142022596 CA9421699 |
218 | T>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA9421696 rs781052184 |
219 | P>L | No |
ClinGen ExAC gnomAD |
|
| rs781052184 | 219 | P>L | No |
ExAC gnomAD |
|
| rs201099777 | 219 | P>S | No |
ExAC gnomAD |
|
|
CA9421697 rs201099777 |
219 | P>S | No |
ClinGen ExAC gnomAD |
|
| rs148082366 | 220 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421695 rs148082366 |
220 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA405729239 rs1298302687 |
223 | D>E | No |
ClinGen gnomAD |
|
| rs1298302687 | 223 | D>E | No | gnomAD | |
| rs550762704 | 224 | C>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs550762704 CA9421694 |
224 | C>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs1385891439 | 224 | C>Y | No | gnomAD | |
|
rs1385891439 CA405729236 |
224 | C>Y | No |
ClinGen gnomAD |
|
|
CA405729213 rs1568395800 |
227 | L>Q | No |
ClinGen Ensembl |
|
| rs1568395800 | 227 | L>Q | No | Ensembl | |
| rs1226763996 | 228 | V>E | No | gnomAD | |
|
CA405729207 rs1226763996 |
228 | V>E | No |
ClinGen gnomAD |
|
| rs758645136 | 229 | K>R | No |
ExAC TOPMed gnomAD |
|
|
rs758645136 CA9421692 |
229 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs749545779 | 231 | D>G | No |
ExAC gnomAD |
|
|
rs749545779 CA9421669 |
231 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA405729191 rs1186107848 |
231 | D>N | No |
ClinGen TOPMed |
|
| rs1186107848 | 231 | D>N | No | TOPMed | |
|
rs749545779 CA405729175 |
231 | D>V | No |
ClinGen ExAC gnomAD |
|
| rs749545779 | 231 | D>V | No |
ExAC gnomAD |
|
|
CA9421668 rs780476499 |
232 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs780476499 | 232 | I>T | No |
ExAC TOPMed gnomAD |
|
|
CA9421666 COSM233368 rs199824317 |
233 | V>I | lung skin [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs199824317 | 233 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA405729158 rs1443845892 |
234 | F>Y | No |
ClinGen gnomAD |
|
| rs1443845892 | 234 | F>Y | No | gnomAD | |
| rs1308975164 | 236 | G>V | No | gnomAD | |
|
CA405729140 rs1308975164 |
236 | G>V | No |
ClinGen gnomAD |
|
|
rs1409624182 CA405729115 |
240 | P>S | No |
ClinGen gnomAD |
|
| rs1409624182 | 240 | P>S | No | gnomAD | |
| rs866816156 | 241 | A>S | No | Ensembl | |
|
CA308151533 rs866816156 |
241 | A>S | No |
ClinGen Ensembl |
|
|
CA9421664 rs201284980 |
241 | A>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs201284980 | 241 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs752345288 CA9421663 |
242 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs752345288 | 242 | R>C | No |
ExAC TOPMed gnomAD |
|
|
CA9421662 rs199730724 |
242 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs199730724 | 242 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1181132093 | 246 | C>* | No | gnomAD | |
|
rs1181132093 CA405729077 |
246 | C>* | No |
ClinGen gnomAD |
|
| rs1471244776 | 246 | C>R | No | gnomAD | |
|
rs1471244776 CA405729080 |
246 | C>R | No |
ClinGen gnomAD |
|
|
CA9421661 rs201927970 |
246 | C>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs201927970 | 246 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs201927970 | 246 | C>Y | No |
ExAC TOPMed gnomAD |
|
|
rs201927970 CA308151531 |
246 | C>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs983778630 | 247 | M>I | No |
TOPMed gnomAD |
|
|
rs983778630 CA308151530 |
247 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1481980295 CA405729072 |
247 | M>R | No |
ClinGen gnomAD |
|
| rs1481980295 | 247 | M>R | No | gnomAD | |
| rs776239943 | 248 | Q>R | No |
ExAC gnomAD |
|
|
rs776239943 CA9421660 |
248 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1441780065 CA405729058 |
249 | S>A | No |
ClinGen gnomAD |
|
| rs1441780065 | 249 | S>A | No | gnomAD | |
| rs1265689698 | 250 | D>G | No | gnomAD | |
|
CA405729038 rs1265689698 |
250 | D>G | No |
ClinGen gnomAD |
|
|
rs1248357660 CA405729021 |
253 | K>Q | No |
ClinGen TOPMed |
|
| rs1248357660 | 253 | K>Q | No | TOPMed | |
|
rs1288349372 CA405728977 |
259 | V>G | No |
ClinGen gnomAD |
|
| rs1288349372 | 259 | V>G | No | gnomAD | |
| rs930042447 | 260 | M>I | No |
TOPMed gnomAD |
|
|
CA308151503 rs930042447 |
260 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1047139983 CA308151504 |
260 | M>L | No |
ClinGen TOPMed |
|
| rs1047139983 | 260 | M>L | No | TOPMed | |
| rs199995434 | 262 | T>I | No |
ExAC TOPMed gnomAD |
|
|
CA9421622 rs199995434 |
262 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs199995434 | 262 | T>N | No |
ExAC TOPMed gnomAD |
|
|
rs199995434 CA9421621 |
262 | T>N | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs974089506 | 264 | L>F | No | Ensembl | |
|
rs974089506 CA308151499 |
264 | L>F | No |
ClinGen Ensembl |
|
| rs1600105502 | 266 | V>G | No | Ensembl | |
|
CA405728936 rs1600105502 |
266 | V>G | No |
ClinGen Ensembl |
|
| rs1036211657 | 271 | S>T | No | TOPMed | |
|
CA308151498 rs1036211657 |
271 | S>T | No |
ClinGen TOPMed |
|
|
CA405728882 rs1294380200 |
274 | S>I | No |
ClinGen TOPMed gnomAD |
|
| rs1294380200 | 274 | S>I | No |
TOPMed gnomAD |
|
| rs1294380200 | 274 | S>N | No |
TOPMed gnomAD |
|
|
rs1294380200 CA405728884 |
274 | S>N | No |
ClinGen TOPMed gnomAD |
|
| rs1391350028 | 275 | K>R | No | TOPMed | |
|
rs1391350028 CA405728875 |
275 | K>R | No |
ClinGen TOPMed |
|
| rs200777538 | 276 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421604 rs200777538 |
276 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs956441512 CA308151491 |
277 | P>L | No |
ClinGen Ensembl |
|
| rs956441512 | 277 | P>L | No | Ensembl | |
|
CA9421602 rs764028671 |
279 | S>P | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs764028671 | 279 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs1410800711 | 281 | P>L | No | TOPMed | |
|
rs1410800711 CA405728828 |
281 | P>L | No |
ClinGen TOPMed |
|
|
CA9421601 rs758284323 |
282 | R>C | No |
ClinGen ExAC gnomAD |
|
| rs758284323 | 282 | R>C | No |
ExAC gnomAD |
|
|
rs752774292 CA9421600 |
282 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs752774292 | 282 | R>H | No |
ExAC TOPMed gnomAD |
|
|
rs752774292 CA405728825 |
282 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs752774292 | 282 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs201431198 | 283 | L>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
CA9421599 rs201431198 |
283 | L>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs759725680 | 288 | E>* | No |
ExAC gnomAD |
|
|
rs759725680 CA405728789 |
288 | E>* | No |
ClinGen ExAC gnomAD |
|
| rs759725680 | 288 | E>K | No |
ExAC gnomAD |
|
|
rs759725680 CA9421596 |
288 | E>K | No |
ClinGen ExAC gnomAD |
|
| rs377489489 | 288 | E>V | No |
ExAC TOPMed gnomAD |
|
|
rs377489489 CA9421595 |
288 | E>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs145405290 | 290 | A>P | No |
ESP TOPMed gnomAD |
|
|
rs145405290 CA405728775 |
290 | A>P | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA308151489 rs145405290 |
290 | A>S | No |
ClinGen ESP TOPMed gnomAD |
|
| rs145405290 | 290 | A>S | No |
ESP TOPMed gnomAD |
|
|
CA405728767 rs1288975955 |
291 | G>D | No |
ClinGen TOPMed |
|
| rs1288975955 | 291 | G>D | No | TOPMed | |
| rs1288975955 | 291 | G>V | No | TOPMed | |
|
CA405728768 rs1288975955 |
291 | G>V | No |
ClinGen TOPMed |
|
|
rs766715586 CA9421594 |
292 | Q>R | No |
ClinGen ExAC gnomAD |
|
| rs766715586 | 292 | Q>R | No |
ExAC gnomAD |
|
| rs766521494 | 293 | S>* | No |
ExAC gnomAD |
|
|
rs766521494 CA405728745 |
293 | S>* | No |
ClinGen ExAC gnomAD |
|
| rs766521494 | 293 | S>L | No |
ExAC gnomAD |
|
|
rs766521494 CA9421575 |
293 | S>L | No |
ClinGen ExAC gnomAD |
|
|
rs1467534171 CA405728747 |
293 | S>P | No |
ClinGen gnomAD |
|
| rs1467534171 | 293 | S>P | No | gnomAD | |
|
rs998580258 CA308151303 |
294 | D>G | No |
ClinGen TOPMed gnomAD |
|
| rs998580258 | 294 | D>G | No |
TOPMed gnomAD |
|
| rs760819880 | 294 | D>N | No |
ExAC TOPMed gnomAD |
|
|
rs760819880 CA9421574 |
294 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs1369524108 | 295 | P>T | No | gnomAD | |
|
rs1369524108 CA405728737 |
295 | P>T | No |
ClinGen gnomAD |
|
|
rs773572964 CA9421573 |
296 | F>S | No |
ClinGen ExAC gnomAD |
|
| rs773572964 | 296 | F>S | No |
ExAC gnomAD |
|
|
CA405728720 rs1442846231 |
297 | L>R | No |
ClinGen gnomAD |
|
| rs1442846231 | 297 | L>R | No | gnomAD | |
|
rs1249919598 CA405728700 |
300 | I>T | No |
ClinGen gnomAD |
|
| rs1249919598 | 300 | I>T | No | gnomAD | |
|
CA405728675 rs201892883 |
304 | G>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs201892883 | 304 | G>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs201892883 | 304 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
|
CA9421570 rs201892883 |
304 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA405728654 rs1211378840 |
307 | M>R | No |
ClinGen gnomAD |
|
| rs1211378840 | 307 | M>R | No | gnomAD | |
|
CA405728647 rs1449894424 |
308 | D>V | No |
ClinGen TOPMed gnomAD |
|
| rs1449894424 | 308 | D>V | No |
TOPMed gnomAD |
|
| rs769428350 | 309 | F>L | No |
ExAC TOPMed gnomAD |
|
|
CA9421569 rs769428350 |
309 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405728622 rs1264779561 |
311 | S>F | No |
ClinGen gnomAD |
|
| rs1264779561 | 311 | S>F | No | gnomAD | |
|
rs1312424575 CA405728615 |
312 | K>N | No |
ClinGen TOPMed |
|
| rs1312424575 | 312 | K>N | No | TOPMed | |
| rs1243604840 | 313 | K>T | No | gnomAD | |
|
rs1243604840 CA405728610 |
313 | K>T | No |
ClinGen gnomAD |
|
|
rs1391765148 CA405728595 |
315 | Y>C | No |
ClinGen gnomAD |
|
| rs1391765148 | 315 | Y>C | No | gnomAD | |
| rs1397338323 | 318 | V>A | No | gnomAD | |
|
CA405728560 rs1397338323 |
318 | V>A | No |
ClinGen gnomAD |
|
| rs199549032 | 318 | V>M | No |
ExAC gnomAD |
|
|
CA9421550 rs199549032 |
318 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs1219342252 CA405728527 |
322 | G>D | No |
ClinGen gnomAD |
|
| rs1219342252 | 322 | G>D | No | gnomAD | |
| rs1012161488 | 322 | G>R | No | Ensembl | |
|
CA308151248 rs1012161488 |
322 | G>R | No |
ClinGen Ensembl |
|
| rs776244453 | 323 | E>K | No |
ExAC TOPMed gnomAD |
|
|
CA9421549 rs776244453 |
323 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1281019237 CA405728514 |
324 | C>S | No |
ClinGen gnomAD |
|
| rs1281019237 | 324 | C>S | No | gnomAD | |
|
rs192567608 CA405728508 |
324 | C>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs192567608 | 324 | C>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1289386259 CA405728503 |
325 | D>G | No |
ClinGen gnomAD |
|
| rs1289386259 | 325 | D>G | No | gnomAD | |
|
CA9421547 rs760353657 |
325 | D>N | No |
ClinGen ExAC gnomAD |
|
| rs760353657 | 325 | D>N | No |
ExAC gnomAD |
|
| rs760353657 | 325 | D>Y | No |
ExAC gnomAD |
|
|
CA405728505 rs760353657 |
325 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
rs772799607 CA9421546 |
326 | Q>R | No |
ClinGen ExAC gnomAD |
|
| rs772799607 | 326 | Q>R | No |
ExAC gnomAD |
|
|
CA405728492 rs1353299956 |
327 | G>S | No |
ClinGen TOPMed gnomAD |
|
| rs1353299956 | 327 | G>S | No |
TOPMed gnomAD |
|
| rs1415160764 | 328 | C>Y | No | gnomAD | |
|
CA405728483 rs1415160764 |
328 | C>Y | No |
ClinGen gnomAD |
|
|
CA9421544 rs747869457 |
329 | L>P | No |
ClinGen ExAC gnomAD |
|
| rs747869457 | 329 | L>P | No |
ExAC gnomAD |
|
|
rs768531597 CA9421542 |
330 | A>D | No |
ClinGen ExAC |
|
| rs768531597 | 330 | A>D | No | ExAC | |
|
CA405728463 rs1412364553 |
332 | A>T | No |
ClinGen gnomAD |
|
| rs1412364553 | 332 | A>T | No | gnomAD | |
| rs1239181675 | 335 | L>F | No | gnomAD | |
|
rs1239181675 CA405728441 |
335 | L>F | No |
ClinGen gnomAD |
|
| rs1231558621 | 336 | G>E | No | TOPMed | |
|
CA405728434 rs1231558621 |
336 | G>E | No |
ClinGen TOPMed |
|
|
CA405728436 rs1386210629 |
336 | G>R | No |
ClinGen Ensembl |
|
| rs1386210629 | 336 | G>R | No | Ensembl | |
|
CA9421541 rs749250869 |
337 | W>C | No |
ClinGen ExAC gnomAD |
|
| rs749250869 | 337 | W>C | No |
ExAC gnomAD |
|
|
rs779911693 CA9421522 |
340 | E>* | No |
ClinGen ExAC gnomAD |
|
| rs779911693 | 340 | E>* | No |
ExAC gnomAD |
|
|
CA9421521 rs769910772 |
340 | E>V | No |
ClinGen ExAC gnomAD |
|
| rs769910772 | 340 | E>V | No |
ExAC gnomAD |
|
|
CA405728386 rs1294098915 |
342 | E>K | No |
ClinGen TOPMed |
|
| rs1294098915 | 342 | E>K | No | TOPMed | |
| rs757518230 | 343 | D>E | No |
ExAC TOPMed gnomAD |
|
|
rs757518230 CA9421518 |
343 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1208488672 CA405728378 |
343 | D>N | No |
ClinGen gnomAD |
|
| rs1208488672 | 343 | D>N | No | gnomAD | |
|
CA9421517 RCV000884149 rs147134890 |
344 | L>I | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs147134890 | 344 | L>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs778261733 | 344 | L>P | No |
ExAC gnomAD |
|
|
rs778261733 CA9421516 |
344 | L>P | No |
ClinGen ExAC gnomAD |
|
| rs202130999 | 345 | V>F | No | Ensembl | |
|
rs202130999 CA308151198 |
345 | V>F | No |
ClinGen Ensembl |
|
|
rs201687831 CA9421512 |
346 | R>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs201687831 | 346 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
CA9421511 rs201687831 |
346 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs201687831 | 346 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
CA9421513 rs199855116 COSM1481025 |
346 | R>W | breast [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
| rs199855116 | 346 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs944648312 | 347 | R>K | No |
TOPMed gnomAD |
|
|
CA308151194 rs944648312 |
347 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1489259283 CA405728355 |
347 | R>S | No |
ClinGen gnomAD |
|
| rs1489259283 | 347 | R>S | No | gnomAD | |
| rs200811137 | 350 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs200811137 CA405728337 |
350 | A>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs200811137 | 350 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
CA9421510 rs200811137 |
350 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs969161300 | 351 | S>G | No | gnomAD | |
|
CA308151190 rs969161300 |
351 | S>G | No |
ClinGen gnomAD |
|
| rs1352190697 | 351 | S>N | No | gnomAD | |
|
CA405728331 rs1352190697 |
351 | S>N | No |
ClinGen gnomAD |
|
|
CA9421509 rs200229086 |
352 | I>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| rs200229086 | 352 | I>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs140902316 | 356 | S>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs140902316 CA9421508 |
356 | S>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs762799591 CA9421506 |
357 | G>E | No |
ClinGen ExAC gnomAD |
|
| rs762799591 | 357 | G>E | No |
ExAC gnomAD |
|
| rs200516283 | 358 | A>G | No |
ExAC TOPMed gnomAD |
|
|
CA9421504 rs200516283 |
358 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs200516283 | 358 | A>V | No |
ExAC TOPMed gnomAD |
|
|
rs200516283 CA9421505 |
358 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs745853916 | 360 | V>F | No |
ExAC gnomAD |
|
|
CA9421501 rs745853916 |
360 | V>F | No |
ClinGen ExAC gnomAD |
|
| rs761994021 | 360 | V>G | No |
ExAC gnomAD |
|
|
CA9421500 rs761994021 |
360 | V>G | No |
ClinGen ExAC gnomAD |
|
|
CA405728248 rs1320069122 |
362 | N>K | No |
ClinGen TOPMed |
|
| rs1320069122 | 362 | N>K | No | TOPMed | |
|
CA308151180 rs201596027 |
363 | P>R | No |
ClinGen gnomAD |
|
| rs201596027 | 363 | P>R | No | gnomAD | |
| rs747221275 | 364 | S>G | No |
ExAC gnomAD |
|
|
rs747221275 CA9421498 |
364 | S>G | No |
ClinGen ExAC gnomAD |
|
| rs1309839331 | 364 | S>N | No | gnomAD | |
|
CA405728234 rs1309839331 |
364 | S>N | No |
ClinGen gnomAD |
|
|
CA405728224 rs1429820547 |
365 | T>I | No |
ClinGen gnomAD |
|
| rs1429820547 | 365 | T>I | No | gnomAD | |
|
CA9421497 rs778208745 |
366 | S>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA405728217 rs778208745 |
366 | S>* | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs778208745 | 366 | S>* | No |
ExAC TOPMed gnomAD |
|
| rs778208745 | 366 | S>* | No |
ExAC TOPMed gnomAD |
|
| rs778208745 | 366 | S>L | No |
ExAC TOPMed gnomAD |
|
|
rs778208745 CA405728216 |
366 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs1167720468 | 368 | S>F | No |
TOPMed gnomAD |
|
|
CA405728198 rs1167720468 |
368 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
CA9421496 rs758700236 |
369 | P>S | No |
ClinGen ExAC gnomAD |
|
| rs758700236 | 369 | P>S | No |
ExAC gnomAD |
|
|
rs1200341993 CA405728180 |
370 | K>N | No |
ClinGen TOPMed |
|
| rs1200341993 | 370 | K>N | No | TOPMed | |
| rs748401916 | 370 | K>R | No |
ExAC gnomAD |
|
|
CA9421495 rs748401916 |
370 | K>R | No |
ClinGen ExAC gnomAD |
|
| rs1490301559 | 372 | S>F | No | gnomAD | |
|
CA405728165 rs1490301559 |
372 | S>F | No |
ClinGen gnomAD |
|
|
rs1600101770 CA405728169 |
372 | S>P | No |
ClinGen Ensembl |
|
| rs1600101770 | 372 | S>P | No | Ensembl | |
|
rs113734329 CA9421494 |
373 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| rs113734329 | 373 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1250784750 CA405728149 |
374 | P>L | No |
ClinGen TOPMed gnomAD |
|
| rs1250784750 | 374 | P>L | No |
TOPMed gnomAD |
|
| rs1250784750 | 374 | P>Q | No |
TOPMed gnomAD |
|
|
CA405728151 rs1250784750 |
374 | P>Q | No |
ClinGen TOPMed gnomAD |
|
| rs1481255336 | 374 | P>T | No | gnomAD | |
|
CA405728154 rs1481255336 |
374 | P>T | No |
ClinGen gnomAD |
|
|
CA405728143 rs1600101712 |
375 | P>S | No |
ClinGen Ensembl |
|
| rs1600101712 | 375 | P>S | No | Ensembl | |
| rs200296263 | 378 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
|
CA405728112 rs200296263 |
378 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs201335739 CA308151172 |
378 | D>N | No |
ClinGen Ensembl |
|
| rs201335739 | 378 | D>N | No | Ensembl | |
| rs756815336 | 379 | E>G | No |
ExAC gnomAD |
|
|
CA9421490 rs756815336 |
379 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA9421491 rs202127838 |
379 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs202127838 | 379 | E>K | No |
ExAC TOPMed gnomAD |
|
|
rs751205777 CA9421489 |
380 | A>S | No |
ClinGen ExAC gnomAD |
|
| rs751205777 | 380 | A>S | No |
ExAC gnomAD |
|
|
rs1374162204 CA405728075 |
383 | T>R | No |
ClinGen TOPMed |
|
| rs1374162204 | 383 | T>R | No | TOPMed | |
|
CA9421487 rs763666706 |
386 | E>V | No |
ClinGen ExAC gnomAD |
|
| rs763666706 | 386 | E>V | No |
ExAC gnomAD |
No associated diseases with Q8IXJ6
1 regional properties for Q8IXJ6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Sirtuin family, catalytic core domain | 57 - 338 | IPR026590 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.1.286 | Transferring groups other than amino-acyl groups |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11085 | NAD-DEPENDENT PROTEIN DEACYLASE SIRTUIN-5, MITOCHONDRIAL-RELATED |
| PANTHER Subfamily | PTHR11085:SF10 | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2 |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
27 GO annotations of cellular component
| Name | Definition |
|---|---|
| centriole | A cellular organelle, found close to the nucleus in many eukaryotic cells, consisting of a small cylinder with microtubular walls, 300-500 nm long and 150-250 nm in diameter. It contains nine short, parallel, peripheral microtubular fibrils, each fibril consisting of one complete microtubule fused to two incomplete microtubules. Cells usually have two centrioles, lying at right angles to each other. At division, each pair of centrioles generates another pair and the twin pairs form the pole of the mitotic spindle. |
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| chromatin silencing complex | Any protein complex that mediates changes in chromatin structure that result in transcriptional silencing. |
| chromosome | A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information. |
| chromosome, telomeric region | The end of a linear chromosome, required for the integrity and maintenance of the end. A chromosome telomere usually includes a region of telomerase-encoded repeats the length of which rarely exceeds 20 bp each and that permits the formation of a telomeric loop (T-loop). The telomeric repeat region is usually preceded by a sub-telomeric region that is gene-poor but rich in repetitive elements. Some telomeres only consist of the latter part (for eg. D. melanogaster telomeres). |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| glial cell projection | A prolongation or process extending from a glial cell. |
| growth cone | The migrating motile tip of a growing neuron projection, where actin accumulates, and the actin cytoskeleton is the most dynamic. |
| heterochromatin | A compact and highly condensed form of chromatin that is refractory to transcription. |
| juxtaparanode region of axon | A region of an axon near a node of Ranvier that is between the paranode and internode regions. |
| lateral loop | Non-compact myelin located adjacent to the nodes of Ranvier in a myelin segment. These non-compact regions include cytoplasm from the cell responsible for synthesizing the myelin. Lateral loops are found in the paranodal region adjacent to the nodes of Ranvier, while Schmidt-Lantermann clefts are analogous structures found within the compact myelin internode. |
| meiotic spindle | A spindle that forms as part of meiosis. Several proteins, such as budding yeast Spo21p, fission yeast Spo2 and Spo13, and C. elegans mei-1, localize specifically to the meiotic spindle and are absent from the mitotic spindle. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| mitotic spindle | A spindle that forms as part of mitosis. Mitotic and meiotic spindles contain distinctive complements of proteins associated with microtubules. |
| myelin sheath | An electrically insulating fatty layer that surrounds the axons of many neurons. It is an outgrowth of glial cells: Schwann cells supply the myelin for peripheral neurons while oligodendrocytes supply it to those of the central nervous system. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| paranodal junction | A highly specialized cell-cell junction found in vertebrates, which forms between a neuron and a glial cell, and has structural similarity to Drosophila septate junctions. It flanks the node of Ranvier in myelinated nerve and electrically isolates the myelinated from unmyelinated nerve segments and physically separates the voltage-gated sodium channels at the node from the cluster of potassium channels underneath the myelin sheath. |
| paranode region of axon | An axon part that is located adjacent to the nodes of Ranvier and surrounded by lateral loop portions of myelin sheath. |
| perikaryon | The portion of the cell soma (neuronal cell body) that excludes the nucleus. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| Schmidt-Lanterman incisure | Regions within compact myelin in which the cytoplasmic faces of the enveloping myelin sheath are not tightly juxtaposed, and include cytoplasm from the cell responsible for making the myelin. Schmidt-Lanterman incisures occur in the compact myelin internode, while lateral loops are analogous structures found in the paranodal region adjacent to the nodes of Ranvier. |
| spindle | The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart. |
14 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| histone acetyltransferase binding | Binding to an histone acetyltransferase. |
| histone deacetylase activity | Catalysis of the reaction: histone N6-acetyl-L-lysine + H2O = histone L-lysine + acetate. This reaction represents the removal of an acetyl group from a histone, a class of proteins complexed to DNA in chromatin and chromosomes. |
| histone deacetylase binding | Binding to histone deacetylase. |
| NAD+ ADP-ribosyltransferase activity | Catalysis of the reaction: NAD+ + (ADP-D-ribosyl)(n)-acceptor = nicotinamide + (ADP-D-ribosyl)(n+1)-acceptor. |
| NAD+ binding | Binding to the oxidized form, NAD, of nicotinamide adenine dinucleotide, a coenzyme involved in many redox and biosynthetic reactions. |
| NAD-dependent histone deacetylase activity | Catalysis of the reaction: histone N6-acetyl-L-lysine + H2O = histone L-lysine + acetate. This reaction requires the presence of NAD, and represents the removal of an acetyl group from a histone. |
| NAD-dependent histone deacetylase activity (H4-K16 specific) | Catalysis of the reaction: histone H4 N6-acetyl-L-lysine (position 16) + H2O = histone H4 L-lysine (position 16) + acetate. This reaction requires the presence of NAD, and represents the removal of an acetyl group from lysine at position 16 of the histone H4 protein. |
| NAD-dependent protein deacetylase activity | Catalysis of the removal of one or more acetyl groups from a protein, requiring NAD. |
| protein deacetylase activity | Catalysis of the reaction: H2O + N6-acetyl-L-lysyl- |
| tubulin deacetylase activity | Catalysis of the reaction: N-acetyl(alpha-tubulin) + H2O = alpha-tubulin + acetate. |
| ubiquitin binding | Binding to ubiquitin, a protein that when covalently bound to other cellular proteins marks them for proteolytic degradation. |
| zinc ion binding | Binding to a zinc ion (Zn). |
55 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| cellular lipid catabolic process | The chemical reactions and pathways resulting in the breakdown of lipids, as carried out by individual cells. |
| cellular response to caloric restriction | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a of caloric restriction, insufficient food energy intake. |
| cellular response to epinephrine stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an epinephrine stimulus. Epinephrine is a catecholamine that has the formula C9H13NO3; it is secreted by the adrenal medulla to act as a hormone, and released by certain neurons to act as a neurotransmitter active in the central nervous system. |
| cellular response to hepatocyte growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hepatocyte growth factor stimulus. |
| cellular response to hypoxia | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension. Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%, results in metabolic adaptation at both the cellular and organismal level. |
| cellular response to molecule of bacterial origin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin. |
| cellular response to oxidative stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of oxidative stress, a state often resulting from exposure to high levels of reactive oxygen species, e.g. superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals. |
| hepatocyte growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a hepatocyte growth factor receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| heterochromatin assembly | An epigenetic gene silencing mechanism in which chromatin is compacted into heterochromatin, resulting in a chromatin conformation refractory to transcription. This process starts with heterochromatin nucleation, its spreading, and ends with heterochromatin boundary formation. |
| histone deacetylation | The modification of histones by removal of acetyl groups. |
| histone H3 deacetylation | The modification of histone H3 by the removal of one or more acetyl groups. |
| histone H4 deacetylation | The modification of histone H4 by the removal of one or more acetyl groups. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| meiotic cell cycle | Progression through the phases of the meiotic cell cycle, in which canonically a cell replicates to produce four offspring with half the chromosomal content of the progenitor cell via two nuclear divisions. |
| mitotic nuclear membrane reassembly | The mitotic cell cycle process involving ESCRTIII that results in reformation of the nuclear envelope after mitotic nuclear division. In organisms undergoing closed mitosis this involves resealing or 'repair' of the nuclear envelope in the nuclear bridge. |
| myelination in peripheral nervous system | The process in which neuronal axons and dendrites become coated with a segmented lipid-rich sheath (myelin) to enable faster and more energetically efficient conduction of electrical impulses. The sheath is formed by the cell membranes of Schwann cells in the peripheral nervous system. Adjacent myelin segments are separated by a non-myelinated stretch of axon called a node of Ranvier. |
| negative regulation of autophagy | Any process that stops, prevents, or reduces the frequency, rate or extent of autophagy. Autophagy is the process in which cells digest parts of their own cytoplasm. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of defense response to bacterium | Any process that stops, prevents or reduces the frequency, rate or extent of defense response to bacterium. |
| negative regulation of fat cell differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of adipocyte differentiation. |
| negative regulation of NLRP3 inflammasome complex assembly | Any process that stops, prevents or reduces the frequency, rate or extent of NLRP3 inflammasome complex assembly. |
| negative regulation of oligodendrocyte progenitor proliferation | Any process that stops or decreases the rate or extent of oligodendrocyte progenitor proliferation. |
| negative regulation of peptidyl-threonine phosphorylation | Any process that decreases the frequency, rate or extent of peptidyl-threonine phosphorylation. Peptidyl-threonine phosphorylation is the phosphorylation of peptidyl-threonine to form peptidyl-O-phospho-L-threonine. |
| negative regulation of protein catabolic process | Any process that stops, prevents or reduces the frequency, rate or extent of protein catabolic process. |
| negative regulation of reactive oxygen species metabolic process | Any process that stops, prevents or reduces the frequency, rate or extent of reactive oxygen species metabolic process. |
| negative regulation of striated muscle tissue development | Any process that stops, prevents, or reduces the frequency, rate or extent of striated muscle development. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| negative regulation of transcription from RNA polymerase II promoter in response to hypoxia | Any process that decreases the frequency, rate or extent of transcription from an RNA polymerase II promoter as a result of a hypoxia stimulus. |
| negative regulation of transcription, DNA-templated | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| peptidyl-lysine deacetylation | The removal of an acetyl group from an acetylated lysine residue in a peptide or protein. |
| phosphatidylinositol 3-kinase signaling | A series of reactions within the signal-receiving cell, mediated by the intracellular phosphatidylinositol 3-kinase (PI3K). Many cell surface receptor linked signaling pathways signal through PI3K to regulate numerous cellular functions. |
| positive regulation of attachment of spindle microtubules to kinetochore | Any process that activates or increases the frequency, rate or extent of the attachment of spindle microtubules to the kinetochore. |
| positive regulation of cell division | Any process that activates or increases the frequency, rate or extent of cell division. |
| positive regulation of DNA binding | Any process that increases the frequency, rate or extent of DNA binding. DNA binding is any process in which a gene product interacts selectively with DNA (deoxyribonucleic acid). |
| positive regulation of execution phase of apoptosis | Any process that activates or increases the frequency, rate or extent of execution phase of apoptosis. |
| positive regulation of meiotic nuclear division | Any process that activates or increases the frequency, rate or extent of meiosis. |
| positive regulation of oocyte maturation | Any process that activates or increases the frequency, rate or extent of oocyte maturation. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia | Any positive regulation of proteasomal ubiquitin-dependent protein catabolic process that is involved in a cellular response to hypoxia. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| proteasome-mediated ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| protein ADP-ribosylation | The transfer, from NAD, of ADP-ribose to protein amino acids. |
| protein deacetylation | The removal of an acetyl group from a protein amino acid. An acetyl group is CH3CO-, derived from acetic |
| protein kinase B signaling | A series of reactions, mediated by the intracellular serine/threonine kinase protein kinase B (also called AKT), which occurs as a result of a single trigger reaction or compound. |
| rDNA heterochromatin assembly | The formation of heterochromatin at ribosomal DNA, characterized by the modified histone H3K9me3. |
| regulation of cell cycle | Any process that modulates the rate or extent of progression through the cell cycle. |
| regulation of exit from mitosis | Any process involved in the progression from anaphase/telophase to G1 that is associated with a conversion from high to low mitotic CDK activity. |
| regulation of myelination | Any process that modulates the frequency, rate or extent of the formation of a myelin sheath around nerve axons. |
| regulation of phosphorylation | Any process that modulates the frequency, rate or extent of addition of phosphate groups into a molecule. |
| response to redox state | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating redox state. Redox state refers to the balance of oxidized versus reduced forms of electron donors and acceptors in an organelle, cell or organ; plastoquinone, glutathione (GSH/GSSG), and nicotinamide nucleotides (NAD+/NADH and NADP+/NADPH) are among the most important. |
| substantia nigra development | The progression of the substantia nigra over time from its initial formation until its mature state. The substantia nigra is the layer of gray substance that separates the posterior parts of the cerebral peduncles (tegmentum mesencephali) from the anterior parts; it normally includes a posterior compact part with many pigmented cells (pars compacta) and an anterior reticular part whose cells contain little pigment (pars reticularis). |
| subtelomeric heterochromatin assembly | The compaction of chromatin into heterochromatin at the subtelomeric region. |
| tubulin deacetylation | The removal of an acetyl group from tubulin. An acetyl group is CH3CO-, derived from acetic |
9 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P53686 | HST2 | NAD-dependent protein deacetylase HST2 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| Q9I7I7 | Sirt2 | NAD-dependent protein deacetylase Sirt2 | Drosophila melanogaster (Fruit fly) | SS |
| Q96EB6 | SIRT1 | NAD-dependent protein deacetylase sirtuin-1 | Homo sapiens (Human) | PR |
| Q8N6T7 | SIRT6 | NAD-dependent protein deacylase sirtuin-6 | Homo sapiens (Human) | PR |
| Q9NTG7 | SIRT3 | NAD-dependent protein deacetylase sirtuin-3, mitochondrial | Homo sapiens (Human) | SS |
| Q8R104 | Sirt3 | NAD-dependent protein deacetylase sirtuin-3 | Mus musculus (Mouse) | SS |
| Q8VDQ8 | Sirt2 | NAD-dependent protein deacetylase sirtuin-2 | Mus musculus (Mouse) | SS |
| Q5RJQ4 | Sirt2 | NAD-dependent protein deacetylase sirtuin-2 | Rattus norvegicus (Rat) | PR |
| Q7ZVK3 | sirt2 | NAD-dependent protein deacetylase sirtuin-2 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAEPDPSHPL | ETQAGKVQEA | QDSDSDSEGG | AAGGEADMDF | LRNLFSQTLS | LGSQKERLLD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ELTLEGVARY | MQSERCRRVI | CLVGAGISTS | AGIPDFRSPS | TGLYDNLEKY | HLPYPEAIFE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ISYFKKHPEP | FFALAKELYP | GQFKPTICHY | FMRLLKDKGL | LLRCYTQNID | TLERIAGLEQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EDLVEAHGTF | YTSHCVSASC | RHEYPLSWMK | EKIFSEVTPK | CEDCQSLVKP | DIVFFGESLP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ARFFSCMQSD | FLKVDLLLVM | GTSLQVQPFA | SLISKAPLST | PRLLINKEKA | GQSDPFLGMI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MGLGGGMDFD | SKKAYRDVAW | LGECDQGCLA | LAELLGWKKE | LEDLVRREHA | SIDAQSGAGV |
| 370 | 380 | ||||
| PNPSTSASPK | KSPPPAKDEA | RTTEREKPQ |