Q8IWV7
Gene name |
UBR1 |
Protein name |
E3 ubiquitin-protein ligase UBR1 |
Names |
EC 2.3.2.27 , N-recognin-1 , RING-type E3 ubiquitin transferase UBR1 , Ubiquitin-protein ligase E3-alpha-1 , Ubiquitin-protein ligase E3-alpha-I |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:197131 |
EC number |
2.3.2.27: Aminoacyltransferases |
Protein Class |
|
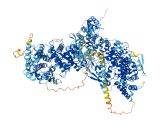
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1-1028 (N-terminal region containing three substrate-binding sites) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Du F et al. (2002) "Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain", Proceedings of the National Academy of Sciences of the United States of America, 99, 14110-5
Autoinhibited structure

Activated structure

3 structures for Q8IWV7
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3NY1 | X-ray | 208 A | A/B | 97-168 | PDB |
| 5TDC | X-ray | 161 A | A/C | 98-168 | PDB |
| AF-Q8IWV7-F1 | Predicted | AlphaFoldDB |
1568 variants for Q8IWV7
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| VAR_075179 | 122 | V>L | JBS; decreased, but detectable activity in a yeast-based assay [UniProt] | Yes | UniProt |
| VAR_075180 | 127 | C>F | JBS [UniProt] | Yes | UniProt |
|
RCV000004942 CA117012 VAR_024741 rs119477054 |
136 | H>R | Johanson-Blizzard syndrome Johanson-blizzard syndrome (jbs) JBS; prevents proper folding of the UBR-type zinc finger; may decrease protein stability; loss of activity in a yeast-based assay [ClinVar, Ensembl, UniProt] | Yes |
ClinGen ClinVar UniProt dbSNP gnomAD |
| VAR_075181 | 166 | H>R | JBS [UniProt] | Yes | UniProt |
|
RCV000497879 CA7518977 rs190360605 RCV001252904 |
202 | V>I | Microcephaly [ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
rs2033815615 VAR_075182 |
217 | L>R | JBS [UniProt] | Yes |
TOPMed UniProt |
| VAR_075183 | 286 | I>R | JBS [UniProt] | Yes | UniProt |
|
rs147396426 RCV001330530 |
316 | R>H | Johanson-Blizzard syndrome [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
| VAR_075184 | 317 | L>P | JBS [UniProt] | Yes | UniProt |
| VAR_075185 | 389 | A>del | JBS [UniProt] | Yes | UniProt |
|
rs77360687 RCV000971625 RCV001819115 RCV001262701 |
405 | S>G | Johanson-Blizzard syndrome [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
VAR_075186 rs1158249054 |
427 | L>R | JBS; uncertain significance [UniProt] | Yes |
UniProt dbSNP gnomAD |
|
CA117013 RCV000004944 rs119477055 |
513 | Q>* | Johanson-Blizzard syndrome Johanson-blizzard syndrome (jbs) [ClinVar, Ensembl] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
RCV000431678 CA7518664 rs768686147 VAR_075187 |
563 | A>D | JBS [UniProt] | Yes |
ClinGen ClinVar UniProt ExAC TOPMed dbSNP gnomAD |
| VAR_075188 | 660 | V>del | JBS [UniProt] | Yes | UniProt |
| VAR_075189 | 700 | S>P | JBS [UniProt] | Yes | UniProt |
|
rs1388367359 VAR_075190 RCV001266507 |
754 | R>C | Inborn genetic diseases JBS [ClinVar, UniProt] | Yes |
ClinVar UniProt dbSNP gnomAD |
|
VAR_075191 RCV001330528 RCV001859277 rs1567131023 |
754 | R>H | Johanson-Blizzard syndrome JBS [ClinVar, UniProt] | Yes |
ClinVar UniProt Ensembl dbSNP |
|
CA392070953 RCV002063857 rs1131691524 RCV000494094 |
758 | G>E | Johanson-Blizzard syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV002058447 RCV002503952 CA7518386 VAR_052116 RCV000253719 rs35069201 |
899 | I>V | Johanson-Blizzard syndrome [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001266518 rs267604210 |
1019 | R>* | Inborn genetic diseases [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
RCV000504515 RCV000224677 CA7518195 rs142285781 COSM1372964 RCV003137831 |
1097 | T>M | Johanson-Blizzard syndrome large_intestine [ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| VAR_075192 | 1102 | Q>E | JBS; strong decrease in activity in a yeast-based assay [UniProt] | Yes | UniProt |
|
RCV000323538 RCV002519152 CA7518158 rs751554684 |
1152 | Y>C | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1235541565 VAR_075193 |
1242 | R>G | JBS [UniProt] | Yes |
UniProt TOPMed dbSNP gnomAD |
| VAR_024742 | 1279 | G>S | JBS [UniProt] | Yes | UniProt |
|
RCV000190636 rs797045112 CA090953 |
1369 | C>* | Johanson-Blizzard syndrome [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
COSM3501336 VAR_075194 |
1426 | P>L | Variant assessed as Somatic; MODERATE impact. JBS [NCI-TCGA, UniProt] | Yes |
NCI-TCGA Cosmic UniProt |
|
RCV002016188 rs1480939799 VAR_075195 |
1427 | S>F | JBS [UniProt] | Yes |
ClinVar UniProt TOPMed dbSNP gnomAD |
|
VAR_075196 rs140972409 |
1431 | S>P | JBS [UniProt] | Yes |
UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
rs374049779 RCV001785111 RCV003560847 |
1508 | Y>* | Johanson-Blizzard syndrome [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
| VAR_075197 | 1661 | G>R | JBS [UniProt] | Yes | UniProt |
|
rs963383651 RCV001170015 |
1673 | C>W | Johanson-Blizzard syndrome [ClinVar] | Yes |
ClinVar TOPMed dbSNP |
|
RCV000895159 RCV001252871 rs141828250 |
1712 | R>H | Microcephaly [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs760804119 RCV002592650 RCV001988699 |
1732 | A>S | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
| rs1451987639 | 2 | A>V | No | gnomAD | |
| rs1370737871 | 3 | D>E | No | gnomAD | |
| COSM4521223 | 4 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1225011799 | 5 | E>* | No | gnomAD | |
| rs754961922 | 6 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs754961922 | 6 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs746895662 | 6 | A>V | No |
ExAC gnomAD |
|
| rs141411839 | 7 | G>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs141411839 | 7 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1273608997 | 9 | T>A | No | gnomAD | |
|
RCV001968655 rs764428187 |
9 | T>I | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs1273608997 | 9 | T>P | No | gnomAD | |
| rs764428187 | 9 | T>S | No |
ExAC gnomAD |
|
| rs375377043 | 10 | E>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs760140949 | 10 | E>D | No |
ExAC gnomAD |
|
| rs375377043 | 10 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs756224722 | 10 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs375377043 | 10 | E>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1204703176 | 11 | R>K | No |
TOPMed gnomAD |
|
| rs2034293410 | 12 | M>L | No | gnomAD | |
| rs1234239450 | 12 | M>T | No | TOPMed | |
| rs774833230 | 13 | E>A | No |
ExAC gnomAD |
|
| rs2034293308 | 14 | I>V | No | gnomAD | |
| rs765927915 | 15 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs1476873906 | 15 | S>I | No | TOPMed | |
| rs76172507 | 16 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs76172507 | 16 | A>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs772637882 | 16 | A>P | No |
ExAC TOPMed gnomAD |
|
|
rs76172507 RCV000886850 |
16 | A>V | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1166673661 | 17 | E>* | No |
TOPMed gnomAD |
|
| rs768302047 | 18 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs150176535 | 20 | Q>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs546134585 | 20 | Q>P | No |
1000Genomes ExAC TOPMed |
|
| rs1262786849 | 21 | T>I | No |
TOPMed gnomAD |
|
| rs1262786849 | 21 | T>N | No |
TOPMed gnomAD |
|
| rs1596147548 | 21 | T>P | No | Ensembl | |
| TCGA novel | 22 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749382782 | 22 | P>L | No |
ExAC gnomAD |
|
| rs768101403 | 24 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs768101403 | 24 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs756384025 | 25 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs911475352 | 26 | A>T | No |
TOPMed gnomAD |
|
| rs1285572810 | 26 | A>V | No | gnomAD | |
| rs1200676814 | 28 | W>L | No | gnomAD | |
| rs1307640526 | 28 | W>R | No |
TOPMed gnomAD |
|
| rs1260647938 | 29 | W>* | No | gnomAD | |
| rs757716545 | 29 | W>R | No |
ExAC gnomAD |
|
| rs1215211413 | 30 | D>G | No | gnomAD | |
|
COSM3987959 rs1387789565 |
30 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs1350797845 | 31 | Q>R | No |
TOPMed gnomAD |
|
| rs528718889 | 33 | V>A | No |
1000Genomes ExAC gnomAD |
|
|
COSM1188989 rs528718889 |
33 | V>D | lung [Cosmic] | No |
cosmic curated 1000Genomes ExAC gnomAD |
| rs1013810338 | 34 | D>H | No |
TOPMed gnomAD |
|
| rs1333958683 | 36 | Y>C | No | TOPMed | |
| rs749436984 | 37 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1028277827 | 38 | A>G | No | TOPMed | |
| rs1311589528 | 38 | A>S | No | gnomAD | |
| rs1028277827 | 38 | A>V | No | TOPMed | |
| rs2034033814 | 39 | F>L | No | Ensembl | |
| rs1283656979 | 40 | L>S | No | gnomAD | |
| rs2034033717 | 41 | H>Y | No |
TOPMed gnomAD |
|
| rs2034033690 | 42 | H>R | No | TOPMed | |
| rs1347570103 | 43 | L>V | No | gnomAD | |
| rs2034033503 | 46 | L>S | No | Ensembl | |
| rs1316782070 | 47 | V>A | No | TOPMed | |
| rs2034033429 | 48 | P>S | No | gnomAD | |
| rs759415631 | 49 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs774125752 | 50 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs766255677 | 52 | F>C | No |
ExAC TOPMed gnomAD |
|
| rs2141360904 | 52 | F>L | No | Ensembl | |
|
COSM358925 rs1177562942 |
55 | M>I | lung [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs2034033256 | 55 | M>T | No | TOPMed | |
| rs1481641383 | 56 | D>V | No | gnomAD | |
| rs2034033074 | 62 | Q>L | No | gnomAD | |
| rs768691069 | 63 | E>D | No |
ExAC gnomAD |
|
| rs2034032946 | 64 | E>K | No |
TOPMed gnomAD |
|
| rs1414479860 | 66 | V>G | No | gnomAD | |
| rs2034032881 | 66 | V>I | No | gnomAD | |
| rs747297205 | 67 | Q>R | No |
ExAC gnomAD |
|
| rs1263709896 | 68 | M>T | No | gnomAD | |
| rs772225984 | 70 | I>V | No |
ExAC TOPMed gnomAD |
|
| COSM4054767 | 71 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1315852077 | 72 | T>A | No | gnomAD | |
| rs140686087 | 72 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs267604211 | 73 | P>L | No | Ensembl | |
| rs267604212 | 73 | P>S | No | Ensembl | |
| rs2034032540 | 74 | L>V | No |
TOPMed gnomAD |
|
| rs779298328 | 75 | E>Q | No |
ExAC gnomAD |
|
| rs1042167924 | 79 | F>L | No |
TOPMed gnomAD |
|
| COSM6142136 | 80 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1159947356 | 82 | D>H | No | TOPMed | |
| rs777191438 | 83 | P>S | No |
ExAC gnomAD |
|
| rs2034032231 | 85 | I>V | No | gnomAD | |
| rs947212743 | 86 | C>S | No |
TOPMed gnomAD |
|
| rs752458307 | 87 | L>F | No |
ExAC TOPMed |
|
| rs1398567348 | 89 | K>T | No | gnomAD | |
| rs2034032066 | 92 | H>R | No | TOPMed | |
| rs754633169 | 93 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1475523602 | 94 | G>R | No |
TOPMed gnomAD |
|
| rs751329469 | 95 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs751329469 | 95 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1374216532 | 98 | L>R | No |
TOPMed gnomAD |
|
| rs202158437 | 101 | R>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1243619104 | 102 | V>I | No |
TOPMed gnomAD |
|
| rs1596137906 | 105 | S>G | No | Ensembl | |
| rs773195975 | 105 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1230952016 | 109 | T>A | No | Ensembl | |
| rs1053447941 | 109 | T>S | No | Ensembl | |
| rs747688821 | 114 | D>G | No |
ExAC gnomAD |
|
| rs1052837637 | 114 | D>N | No | Ensembl | |
| rs140294470 | 117 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2141357793 | 121 | C>G | No | Ensembl | |
| COSM1372968 | 122 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM6142137 | 124 | C>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs751192063 | 125 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs2033986642 | 126 | D>N | No | gnomAD | |
| rs904194823 | 130 | D>N | No | Ensembl | |
| rs1596136293 | 132 | V>I | No | Ensembl | |
| rs576341424 | 135 | N>S | No |
1000Genomes gnomAD |
|
| rs119477054 | 136 | H>L | Johanson-blizzard syndrome (jbs) [Ensembl] | No | gnomAD |
| rs765166092 | 137 | R>C | No |
ExAC gnomAD |
|
|
RCV000936636 rs201509699 |
137 | R>H | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs2033986200 | 139 | K>R | No | TOPMed | |
| rs757922598 | 140 | M>I | No |
ExAC gnomAD |
|
| rs1290564440 | 141 | H>P | No |
TOPMed gnomAD |
|
| rs1290564440 | 141 | H>R | No |
TOPMed gnomAD |
|
| rs983350121 | 141 | H>Y | No | TOPMed | |
| rs2033871171 | 144 | T>I | No | TOPMed | |
| rs59574876 | 145 | G>* | No | Ensembl | |
| rs2033871055 | 148 | F>S | No | TOPMed | |
| rs2033871055 | 148 | F>Y | No | TOPMed | |
| rs750251744 | 150 | D>A | No |
ExAC gnomAD |
|
| rs778630364 | 154 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs778630364 | 154 | T>P | No |
ExAC TOPMed gnomAD |
|
| COSM2187887 | 157 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033870791 | 158 | K>I | No | TOPMed | |
| COSM3420309 | 158 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs926026005 | 160 | G>C | No | TOPMed | |
| rs950562532 | 160 | G>D | No | Ensembl | |
| rs926026005 | 160 | G>R | No | TOPMed | |
| rs2033870688 | 161 | P>L | No |
TOPMed gnomAD |
|
| TCGA novel | 163 | C>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1208176018 | 163 | C>W | No |
TOPMed gnomAD |
|
| rs144415580 | 164 | V>I | No |
ESP ExAC gnomAD |
|
| rs551627937 | 165 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 166 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1395983775 | 168 | P>R | No | gnomAD | |
| rs759651076 | 169 | G>V | No |
ExAC gnomAD |
|
| rs2033870373 | 170 | R>G | No | Ensembl | |
| rs1216649445 | 171 | A>V | No | gnomAD | |
| rs763034473 | 174 | I>M | No | ExAC | |
| rs766653121 | 174 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs773755976 | 175 | K>E | No |
ExAC gnomAD |
|
| rs139723559 | 179 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs376029932 | 179 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs777067165 COSM1708068 |
181 | P>L | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1384967387 | 182 | L>F | No | gnomAD | |
| rs1000411391 | 183 | N>H | No | Ensembl | |
| rs761040799 | 185 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1474560079 | 188 | V>I | No |
TOPMed gnomAD |
|
| rs775115817 | 189 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs771451861 | 190 | A>T | No |
ExAC gnomAD |
|
| rs2033816759 | 190 | A>V | No | TOPMed | |
| rs2033816726 | 191 | R>K | No | gnomAD | |
| rs745591226 | 192 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1420320653 | 193 | I>M | No | TOPMed | |
| rs1411664896 | 194 | F>L | No |
TOPMed gnomAD |
|
| rs1165416575 | 195 | P>S | No | Ensembl | |
|
rs2033816459 COSM1269655 |
196 | S>L | oesophagus [Cosmic] | No |
cosmic curated Ensembl |
| COSM3501347 | 197 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1332420217 | 198 | I>M | No |
TOPMed gnomAD |
|
| rs756124027 | 198 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs149864087 | 200 | Y>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs2033816062 | 202 | V>A | No | gnomAD | |
| rs985792161 | 206 | I>L | No |
TOPMed gnomAD |
|
| rs985792161 | 206 | I>V | No |
TOPMed gnomAD |
|
| rs1290370381 | 208 | E>D | No | gnomAD | |
| rs1246127178 | 209 | E>K | No | gnomAD | |
|
COSM3816111 rs1361453557 |
210 | E>K | breast [Cosmic] | No |
cosmic curated gnomAD |
| COSM700540 | 212 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs951788556 | 212 | E>Q | No |
TOPMed gnomAD |
|
| rs1222883490 | 212 | E>V | No |
TOPMed gnomAD |
|
| rs752329091 | 215 | P>L | No |
TOPMed gnomAD |
|
| rs752329091 | 215 | P>R | No |
TOPMed gnomAD |
|
| rs62020750 | 215 | P>T | No | Ensembl | |
| COSM3887001 | 216 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs139686505 RCV000957684 |
218 | Q>H | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1364003992 | 218 | Q>R | No | gnomAD | |
| rs1425420064 | 219 | I>K | No | gnomAD | |
| rs2033815310 | 219 | I>M | No | Ensembl | |
| rs1262458487 | 221 | E>V | No | gnomAD | |
| rs1322561177 | 222 | K>E | No | gnomAD | |
| rs1283759042 | 223 | N>S | No | gnomAD | |
| rs1224339094 | 224 | E>K | No | gnomAD | |
| rs1011385263 | 225 | R>S | No | TOPMed | |
| COSM1301139 | 225 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
CA16607782 RCV000434346 rs1057523835 |
226 | Y>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs1381752228 | 227 | Y>C | No | gnomAD | |
| rs1194845467 | 228 | C>F | No | Ensembl | |
| rs778267578 | 230 | L>P | No |
ExAC gnomAD |
|
| rs2033775560 | 232 | N>D | No | TOPMed | |
| TCGA novel | 233 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1757124370 | 234 | E>K | No | TOPMed | |
| rs895218250 | 235 | H>R | No | gnomAD | |
| rs1177677093 | 236 | H>N | No | gnomAD | |
| rs768020434 | 240 | H>D | No |
ExAC gnomAD |
|
| rs751115743 | 241 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs765839529 | 242 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1410206314 | 245 | L>P | No | TOPMed | |
| rs1171876647 | 246 | Q>R | No | TOPMed | |
| COSM470664 | 250 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs150617078 | 251 | C>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs1348809716 | 251 | C>R | No |
TOPMed gnomAD |
|
| rs1348809716 | 251 | C>S | No |
TOPMed gnomAD |
|
| rs2033774730 | 253 | L>R | No | TOPMed | |
| rs746922418 | 254 | A>S | No |
ExAC TOPMed gnomAD |
|
|
RCV001933678 rs746922418 |
254 | A>T | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs779175153 | 254 | A>V | No |
ExAC gnomAD |
|
| COSM4738804 | 255 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201386354 | 256 | A>S | No | 1000Genomes | |
| rs2033774496 | 257 | Q>R | No | Ensembl | |
| rs2033774387 | 258 | L>S | No | TOPMed | |
| rs569992095 | 258 | L>V | No |
1000Genomes TOPMed gnomAD |
|
| rs2033774387 | 258 | L>W | No | TOPMed | |
| COSM3816109 | 261 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1382765470 | 261 | T>I | No |
TOPMed gnomAD |
|
| COSM3501346 | 262 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1337770717 | 265 | K>I | No |
TOPMed gnomAD |
|
| rs1337770717 | 265 | K>R | No |
TOPMed gnomAD |
|
| rs1398191306 | 266 | E>K | No |
TOPMed gnomAD |
|
| rs1398191306 | 266 | E>Q | No |
TOPMed gnomAD |
|
| rs775556946 | 267 | G>C | No |
ExAC gnomAD |
|
| rs771991628 | 267 | G>D | No |
ExAC gnomAD |
|
| rs143887505 | 268 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs143887505 | 268 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs200054312 | 269 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs773307468 | 269 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1254286891 | 270 | A>S | No | gnomAD | |
| rs1567139817 | 271 | V>I | No | Ensembl | |
| rs191658926 | 273 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 279 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1230510388 | 280 | Q>R | No | gnomAD | |
| rs755319945 | 281 | E>G | No |
ExAC gnomAD |
|
| rs1279302999 | 282 | A>T | No |
TOPMed gnomAD |
|
| rs1250604636 | 283 | K>E | No | TOPMed | |
|
RCV002524643 RCV000413619 CA7518905 rs747313563 |
284 | E>Q | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs780542374 | 285 | D>G | No |
ExAC gnomAD |
|
| rs758895551 | 287 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1426173252 | 288 | S>G | No | gnomAD | |
| rs753342743 | 288 | S>N | No |
ExAC gnomAD |
|
| rs777575882 | 289 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1025568625 | 293 | V>D | No | TOPMed | |
| rs1025568625 | 293 | V>G | No | TOPMed | |
| rs947395063 | 293 | V>I | No |
TOPMed gnomAD |
|
| rs1013757336 | 295 | Q>K | No |
TOPMed gnomAD |
|
| rs913205469 | 296 | H>R | No |
TOPMed gnomAD |
|
| rs1670162250 | 296 | H>Y | No | Ensembl | |
| rs1167937247 | 299 | H>R | No | gnomAD | |
| COSM3690403 | 302 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1280325469 | 304 | H>Q | No |
TOPMed gnomAD |
|
| rs759455243 | 306 | E>D | No |
ExAC gnomAD |
|
| rs1338190488 | 307 | I>T | No | gnomAD | |
| rs751496698 | 308 | M>V | No |
ExAC gnomAD |
|
| rs1475392062 | 309 | A>T | No | gnomAD | |
| rs766168695 | 310 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs2141337418 | 311 | Q>* | No | 1000Genomes | |
| TCGA novel | 312 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 313 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761882298 | 315 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1190528436 | 316 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2033660442 | 317 | L>H | No | TOPMed | |
| COSM3887000 | 319 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1453129666 | 322 | N>D | No | gnomAD | |
| rs375802608 | 324 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| COSM961879 | 324 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs374589988 | 325 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1319547822 | 325 | M>L | No | Ensembl | |
| rs2033660247 | 325 | M>T | No | Ensembl | |
| rs1283912799 | 326 | S>N | No |
TOPMed gnomAD |
|
| rs1446472380 | 326 | S>R | No |
TOPMed gnomAD |
|
| rs572477955 | 329 | S>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2033653093 | 329 | S>R | No |
TOPMed gnomAD |
|
| rs368606031 | 331 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs759951032 | 331 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs771591555 | 332 | R>K | No |
ExAC gnomAD |
|
| rs1350052903 | 333 | Q>H | No | gnomAD | |
| TCGA novel | 334 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2033652816 | 336 | C>G | No | Ensembl | |
| TCGA novel | 338 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749794424 | 339 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs2033652751 | 339 | C>Y | No | Ensembl | |
| rs2033652682 | 342 | E>D | No |
TOPMed gnomAD |
|
| rs549414094 | 342 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs1344105235 | 343 | E>G | No | TOPMed | |
| rs769412017 | 344 | P>S | No |
ExAC gnomAD |
|
| rs2033652438 | 345 | D>E | No | Ensembl | |
| rs747551403 | 345 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs201544896 | 346 | S>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs775293222 | 347 | E>K | No |
ExAC gnomAD |
|
| rs1183889649 | 348 | N>D | No | gnomAD | |
| rs374557254 | 349 | P>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs764216862 | 352 | I>L | No |
ExAC gnomAD |
|
| rs764216862 | 352 | I>V | No |
ExAC gnomAD |
|
|
rs142798632 RCV001959899 |
355 | L>F | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2033651977 | 356 | M>T | No |
TOPMed gnomAD |
|
| TCGA novel | 358 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1283595232 | 360 | A>S | No | TOPMed | |
| rs759631135 | 360 | A>V | No |
ExAC gnomAD |
|
| rs2033651800 | 361 | K>R | No | TOPMed | |
| rs1329611189 | 362 | L>F | No | gnomAD | |
| rs548089634 | 363 | Y>* | No | 1000Genomes | |
| rs774831690 | 363 | Y>C | No |
ExAC gnomAD |
|
| COSM3501345 | 363 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1230452785 | 365 | G>S | No | gnomAD | |
|
rs755272230 COSM141450 |
367 | R>C | upper_aerodigestive_tract [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs985079300 | 367 | R>H | No |
TOPMed gnomAD |
|
| rs985079300 | 367 | R>L | No |
TOPMed gnomAD |
|
| rs766647532 | 368 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1355677867 | 370 | L>F | No | TOPMed | |
| rs1355677867 | 370 | L>I | No | TOPMed | |
| rs2141336083 | 370 | L>P | No | Ensembl | |
| rs2033643546 | 373 | L>F | No | TOPMed | |
| rs763411888 | 376 | S>T | No |
ExAC gnomAD |
|
| COSM2187873 | 379 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033643492 | 380 | M>I | No | TOPMed | |
| rs930861312 | 380 | M>V | No |
TOPMed gnomAD |
|
| TCGA novel | 380 | M>Y | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1017645736 | 382 | M>I | No | Ensembl | |
|
RCV001870536 rs1275090232 |
382 | M>V | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs2033643419 | 384 | Y>C | No | TOPMed | |
| TCGA novel | 386 | K>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1449976909 | 387 | L>F | No | gnomAD | |
| rs765865717 | 388 | F>L | No |
ExAC gnomAD |
|
| rs773668093 | 388 | F>V | No |
ExAC gnomAD |
|
| rs200801495 | 389 | A>P | No |
TOPMed gnomAD |
|
| rs200801495 | 389 | A>S | No |
TOPMed gnomAD |
|
|
rs200801495 RCV002047736 |
389 | A>T | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs952000855 | 390 | M>L | No |
TOPMed gnomAD |
|
| rs2033643101 | 390 | M>T | No | TOPMed | |
| rs952000855 | 390 | M>V | No |
TOPMed gnomAD |
|
| rs1258310183 | 392 | F>S | No | Ensembl | |
| rs570440171 | 393 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs570440171 RCV002028905 |
393 | V>M | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs768046369 | 394 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs760109232 | 395 | Y>C | No |
ExAC gnomAD |
|
| rs764604121 | 395 | Y>H | No |
ExAC gnomAD |
|
| rs2033620520 | 396 | Y>* | No | Ensembl | |
| rs2033620480 | 398 | Q>* | No | Ensembl | |
| COSM3816108 | 402 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs771737067 | 403 | Y>C | No |
ExAC gnomAD |
|
| TCGA novel | 407 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 408 | H>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770631803 | 408 | H>Y | No |
ExAC gnomAD |
|
| rs2033620193 | 409 | D>G | No | Ensembl | |
| rs371156658 | 410 | R>* | No |
ESP ExAC TOPMed gnomAD |
|
| COSM3370171 | 410 | R>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs942523230 | 411 | S>G | No | gnomAD | |
| rs2033620040 | 411 | S>N | No | gnomAD | |
| rs777572724 | 412 | I>V | No |
ExAC gnomAD |
|
| rs2033619959 | 415 | T>I | No | TOPMed | |
| rs1404676554 | 416 | A>V | No |
TOPMed gnomAD |
|
|
rs756388965 COSM3936758 |
418 | S>* | oesophagus [Cosmic] | No |
cosmic curated Ensembl |
| rs756388965 | 418 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| COSM6077119 | 419 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs786205492 | 420 | Q>E | No | TOPMed | |
|
RCV000171234 CA235927 rs786205492 |
420 | Q>K | No |
ClinGen ClinVar TOPMed dbSNP |
|
| rs1391393903 | 421 | M>T | No | gnomAD | |
| rs1228851998 | 421 | M>V | No |
TOPMed gnomAD |
|
| rs2141334402 | 423 | T>I | No | Ensembl | |
| rs1012412925 | 423 | T>P | No |
TOPMed gnomAD |
|
| rs1164739631 | 424 | V>I | No | gnomAD | |
| rs1164739631 | 424 | V>L | No | gnomAD | |
| rs769727145 | 425 | P>R | No |
ExAC gnomAD |
|
| TCGA novel | 426 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs895175651 | 427 | L>M | No |
TOPMed gnomAD |
|
| rs1162709779 | 429 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs145247056 | 429 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
|
RCV002009275 rs930246370 |
430 | H>Y | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1414492122 | 432 | I>F | No | gnomAD | |
|
CA16042870 RCV000414257 rs1057518231 |
432 | I>T | No |
ClinGen ClinVar TOPMed dbSNP |
|
| rs1477740286 | 435 | Q>* | No | gnomAD | |
| rs2033597900 | 438 | I>L | No | TOPMed | |
| rs770563187 | 438 | I>T | No |
ExAC gnomAD |
|
| rs2033597900 | 438 | I>V | No | TOPMed | |
| rs2033597753 | 441 | I>V | No | TOPMed | |
| rs918826999 | 442 | T>I | No | Ensembl | |
| rs925511677 | 446 | L>R | No | TOPMed | |
| TCGA novel | 447 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM433888 | 451 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1298465458 | 451 | E>G | No | TOPMed | |
| rs1209775298 | 452 | Y>C | No | gnomAD | |
| rs1567138168 | 453 | L>F | No | Ensembl | |
| rs775777743 | 454 | D>H | No |
ExAC gnomAD |
|
| rs772264230 | 455 | R>G | No |
ExAC gnomAD |
|
|
RCV000500201 rs1555448026 CA392072293 |
457 | N>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
| rs1318570816 | 458 | K>R | No |
TOPMed gnomAD |
|
| COSM961877 | 458 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs779007025 | 460 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs779007025 | 460 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs779007025 | 460 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs2033596870 | 462 | Q>* | No |
TOPMed gnomAD |
|
| TCGA novel | 462 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757462259 | 462 | Q>R | No |
ExAC gnomAD |
|
| rs749638136 | 463 | G>D | No |
ExAC gnomAD |
|
| rs1364432490 | 463 | G>S | No | gnomAD | |
| rs778167157 | 466 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs756660723 | 467 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs146690613 | 469 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs767077535 | 472 | V>A | No |
ExAC gnomAD |
|
| rs955425996 | 477 | C>R | No | gnomAD | |
| rs144700609 | 478 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs374446394 | 480 | K>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs751074083 | 481 | Y>C | No |
ExAC gnomAD |
|
| rs751074083 | 481 | Y>F | No |
ExAC gnomAD |
|
| rs1482447945 | 481 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1207178464 | 484 | I>M | No |
TOPMed gnomAD |
|
| rs1178618446 | 485 | S>G | No |
TOPMed gnomAD |
|
| rs1319910016 | 485 | S>R | No | gnomAD | |
| rs2033512963 | 485 | S>T | No | TOPMed | |
| rs2033512840 | 492 | E>G | No | TOPMed | |
| rs765893686 | 493 | R>I | No |
ExAC gnomAD |
|
| rs375204571 | 495 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2033512610 | 496 | M>T | No | Ensembl | |
| rs764918267 | 499 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1393339623 | 501 | G>S | No | gnomAD | |
| rs1432144030 | 502 | F>C | No | gnomAD | |
|
COSM2187863 rs764158113 |
503 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs774341055 | 503 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs774341055 | 503 | R>P | No |
ExAC TOPMed gnomAD |
|
|
rs774341055 COSM170963 RCV002037477 |
503 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs771027157 | 507 | K>R | No |
ExAC gnomAD |
|
| rs1415432663 | 510 | T>A | No | gnomAD | |
| rs763148886 | 510 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs2033511992 | 512 | M>I | No | TOPMed | |
| rs2033497768 | 515 | M>I | No | TOPMed | |
| rs375176883 | 515 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2033497671 | 518 | I>T | No | TOPMed | |
| COSM416930 | 519 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM169612 rs777041521 |
519 | R>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs777041521 | 519 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1290254967 | 522 | V>A | No |
TOPMed gnomAD |
|
| rs769063596 | 526 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs991855606 | 527 | E>D | No | TOPMed | |
| rs2033497350 | 529 | D>Y | No | TOPMed | |
|
COSM3501344 rs2033497309 |
530 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1376546121 | 536 | I>N | No |
TOPMed gnomAD |
|
| rs1176217050 | 538 | I>M | No | gnomAD | |
| rs1363218239 | 538 | I>T | No |
TOPMed gnomAD |
|
| rs2141326351 | 540 | M>I | No | Ensembl | |
| rs747271855 | 540 | M>K | No |
ExAC gnomAD |
|
| rs1455493956 | 540 | M>V | No | gnomAD | |
| rs2141326340 | 543 | K>M | No | 1000Genomes | |
| TCGA novel | 543 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780625695 | 544 | N>K | No |
ExAC gnomAD |
|
| rs970426443 | 545 | I>S | No | Ensembl | |
|
rs1249718415 COSM961876 |
546 | L>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs1249718415 | 546 | L>S | No | gnomAD | |
| rs1207112559 | 547 | L>P | No | gnomAD | |
| rs1316232668 | 548 | M>I | No | TOPMed | |
| COSM5888268 | 550 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033496606 | 552 | W>R | No | Ensembl | |
| rs1025896477 | 554 | A>V | No | Ensembl | |
| rs772411595 | 555 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs560253904 | 556 | D>E | No |
1000Genomes ExAC gnomAD |
|
| rs2033443763 | 557 | E>K | No | Ensembl | |
| rs1567135068 | 559 | L>F | No | Ensembl | |
| TCGA novel | 560 | L>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2033443686 | 560 | L>V | No | Ensembl | |
| rs977997178 | 561 | L>I | No | TOPMed | |
| rs2033443586 | 562 | V>L | No | TOPMed | |
| rs1384495182 | 563 | A>S | No | TOPMed | |
| rs748002347 | 565 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1420060057 | 566 | E>D | No | gnomAD | |
|
TCGA novel rs2033443204 |
569 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs2033443065 | 573 | R>S | No | TOPMed | |
| rs1306913065 | 573 | R>T | No |
TOPMed gnomAD |
|
| rs780831630 | 574 | C>Y | No |
ExAC gnomAD |
|
| rs2141322887 | 575 | S>G | No | Ensembl | |
| rs1398589711 | 575 | S>N | No |
TOPMed gnomAD |
|
| rs1398589711 | 575 | S>T | No |
TOPMed gnomAD |
|
| rs140662099 | 577 | S>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1368900931 | 578 | F>S | No | gnomAD | |
| rs2033442763 | 579 | I>M | No | gnomAD | |
| rs1188920595 | 580 | S>A | No |
TOPMed gnomAD |
|
| rs1188920595 | 580 | S>P | No |
TOPMed gnomAD |
|
| rs568824492 | 581 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs757344571 | 582 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs954696582 | 582 | S>R | No | Ensembl | |
| rs998817919 | 583 | K>E | No |
TOPMed gnomAD |
|
| rs753867982 | 584 | T>I | No |
ExAC gnomAD |
|
| rs753867982 | 584 | T>R | No |
ExAC gnomAD |
|
| rs953401345 | 585 | V>A | No |
TOPMed gnomAD |
|
| rs987679003 | 585 | V>I | No |
TOPMed gnomAD |
|
| rs987679003 | 585 | V>L | No |
TOPMed gnomAD |
|
| rs1356070205 | 588 | S>L | No |
TOPMed gnomAD |
|
| rs775971046 | 591 | H>R | No |
ExAC TOPMed gnomAD |
|
| COSM1372965 | 593 | L>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs148797320 | 595 | T>I | No |
ESP TOPMed |
|
| rs148797320 | 595 | T>R | No |
ESP TOPMed |
|
|
VAR_034467 RCV000960793 rs34568456 |
596 | K>M | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs34568456 | 596 | K>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2033441929 | 597 | S>Y | No | Ensembl | |
| rs2033441837 | 600 | V>I | No | TOPMed | |
| rs1337405557 | 601 | S>P | No | gnomAD | |
|
RCV001867676 rs747081057 |
604 | L>I | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs2033441568 | 605 | V>A | No |
TOPMed gnomAD |
|
| rs2033441607 | 605 | V>I | No | TOPMed | |
| rs771505561 | 606 | S>N | No | ExAC | |
| rs894276290 | 607 | I>M | No | gnomAD | |
| COSM5383341 | 610 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033441181 | 616 | A>V | No | TOPMed | |
| TCGA novel | 617 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764259098 | 617 | G>V | No | Ensembl | |
| rs752857150 | 619 | H>P | No |
ExAC TOPMed gnomAD |
|
| rs752857150 | 619 | H>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 620 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1396219082 | 620 | V>G | No | gnomAD | |
| rs767710329 | 621 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs759990188 | 621 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs767020641 | 624 | R>G | No |
ExAC gnomAD |
|
| rs2033362652 | 626 | G>A | No |
TOPMed gnomAD |
|
| rs1275835930 | 627 | A>G | No |
TOPMed gnomAD |
|
| rs375687713 | 629 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed |
| rs375687713 | 629 | S>L | No |
ESP TOPMed |
|
| rs1199421175 | 630 | R>T | No |
TOPMed gnomAD |
|
| rs573944269 | 632 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs769186907 | 632 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs917772057 | 636 | S>T | No | Ensembl | |
| rs200515587 | 637 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1431721911 | 637 | F>S | No | gnomAD | |
| rs768249377 | 637 | F>V | No |
ExAC gnomAD |
|
| rs1188339117 | 639 | D>G | No | gnomAD | |
| rs2033357842 | 639 | D>N | No | TOPMed | |
| rs761253544 | 640 | F>L | No |
ExAC gnomAD |
|
| rs776169122 | 640 | F>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 641 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs530442432 | 642 | V>A | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 642 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1052567492 | 643 | E>G | No | Ensembl | |
| TCGA novel | 644 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374574697 | 646 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs2033357449 | 646 | V>M | No | TOPMed | |
| TCGA novel | 647 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1482663363 | 648 | Y>C | No | gnomAD | |
| rs1786398861 | 649 | P>S | No | Ensembl | |
| rs1233388497 | 650 | L>V | No | gnomAD | |
|
COSM5849200 rs1313170537 |
651 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1358896992 | 652 | C>Y | No | TOPMed | |
| rs1206613222 | 655 | L>V | No | gnomAD | |
| rs1480711371 | 656 | V>L | No | gnomAD | |
| rs2033356972 | 658 | Q>* | No |
TOPMed gnomAD |
|
| rs2033356972 | 658 | Q>E | No |
TOPMed gnomAD |
|
| rs762423462 | 659 | V>I | No |
TOPMed gnomAD |
|
| rs1232881132 | 660 | V>I | No | gnomAD | |
| rs1335754750 | 663 | M>I | No | gnomAD | |
| rs2033356814 | 663 | M>L | No | Ensembl | |
| rs778728550 | 664 | W>* | No |
ExAC gnomAD |
|
|
COSM1240599 rs1292700460 |
665 | R>* | oesophagus [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1315598396 | 665 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1370426381 | 666 | R>K | No | Ensembl | |
| rs1408086291 | 668 | G>V | No | gnomAD | |
| COSM4853808 | 671 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM961875 | 673 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1465075993 | 673 | S>N | No |
TOPMed gnomAD |
|
| rs770849248 | 673 | S>R | No |
ExAC gnomAD |
|
| rs2033338660 | 675 | V>G | No | TOPMed | |
| rs1181201299 | 675 | V>M | No |
TOPMed gnomAD |
|
| rs1193968504 | 677 | Y>H | No | gnomAD | |
| rs749223205 | 678 | Y>H | No |
ExAC gnomAD |
|
| rs2033338489 | 679 | Q>* | No | gnomAD | |
| TCGA novel | 679 | Q>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs377090547 | 680 | D>A | No |
ESP ExAC TOPMed |
|
| rs377090547 | 680 | D>V | No |
ESP ExAC TOPMed |
|
| TCGA novel | 680 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1171074016 | 683 | C>R | No |
TOPMed gnomAD |
|
| rs2141316869 | 683 | C>Y | No | Ensembl | |
| rs2141316864 | 684 | R>G | No | Ensembl | |
| TCGA novel | 685 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758608456 | 687 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs746259965 | 688 | Y>C | No |
ExAC gnomAD |
|
| COSM961874 | 691 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1235759223 | 692 | I>T | No | gnomAD | |
| rs559405167 | 692 | I>V | No |
1000Genomes ExAC gnomAD |
|
| rs1415129792 | 694 | M>I | No |
TOPMed gnomAD |
|
| COSM961873 | 695 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM6142138 | 696 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033333925 | 697 | I>M | No | TOPMed | |
| rs1167039129 | 698 | G>A | No | gnomAD | |
| rs1462316636 | 699 | A>T | No | gnomAD | |
| rs1172765012 | 702 | M>T | No | gnomAD | |
| TCGA novel | 703 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs145364227 | 705 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 706 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 706 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1256837340 | 707 | F>L | No | gnomAD | |
| rs2033333608 | 708 | L>S | No | TOPMed | |
| rs900434077 | 712 | L>P | No | Ensembl | |
| rs2033333435 | 714 | R>G | No | Ensembl | |
| rs2141316597 | 715 | Y>F | No | Ensembl | |
| rs1250271053 | 716 | E>K | No | gnomAD | |
| rs1020057044 | 716 | E>V | No | TOPMed | |
| rs144190668 | 718 | A>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs144190668 | 718 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1242129337 | 719 | E>D | No | gnomAD | |
| rs1253488499 | 719 | E>K | No |
TOPMed gnomAD |
|
| rs2033333117 | 720 | A>V | No | Ensembl | |
| rs2033333016 | 722 | N>K | No | Ensembl | |
| rs2033332967 | 723 | K>N | No | gnomAD | |
| rs749595417 | 724 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs756572767 | 725 | I>M | No |
ExAC gnomAD |
|
| rs778274546 | 725 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs147478213 | 727 | T>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2033332669 | 728 | K>I | No | Ensembl | |
| rs1391960991 | 730 | Q>R | No | gnomAD | |
| rs2033271252 | 732 | L>V | No | gnomAD | |
| rs778076371 | 733 | I>S | No |
ExAC gnomAD |
|
| rs1328285200 | 736 | Y>C | No |
TOPMed gnomAD |
|
| rs1470773522 | 736 | Y>H | No | gnomAD | |
| rs1192079463 | 737 | N>S | No | gnomAD | |
| rs1454096368 | 738 | T>A | No |
TOPMed gnomAD |
|
| rs2033270902 | 738 | T>I | No | TOPMed | |
| rs574978570 | 739 | L>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1284151275 | 743 | M>I | No | TOPMed | |
| rs2033270571 | 746 | V>D | No |
TOPMed gnomAD |
|
| rs200810218 | 746 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1352680036 | 747 | L>F | No | gnomAD | |
| rs2033270436 | 748 | I>M | No | gnomAD | |
| rs750293170 | 749 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs750293170 | 749 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs1371425274 | 751 | V>M | No | gnomAD | |
| rs2033234492 | 753 | E>A | No | Ensembl | |
| rs1337625465 | 755 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2068241882 | 756 | V>I | No | Ensembl | |
| rs900552063 | 758 | G>R | No |
TOPMed gnomAD |
|
| rs111232529 | 761 | N>H | No | Ensembl | |
| TCGA novel | 765 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 767 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753525736 | 767 | V>I | No |
ExAC gnomAD |
|
| rs200146543 | 768 | T>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 769 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1276984540 | 769 | M>V | No | TOPMed | |
| rs2033233693 | 771 | E>D | No |
TOPMed gnomAD |
|
| rs369213575 | 772 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV001998695 rs2141310474 |
772 | I>V | No |
ClinVar Ensembl dbSNP |
|
| rs2033233542 | 773 | I>M | No |
TOPMed gnomAD |
|
| rs763224715 | 778 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs766428995 | 778 | I>V | No |
ExAC gnomAD |
|
|
rs375913511 COSM470663 |
782 | P>S | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs372946259 | 785 | A>D | No |
ESP TOPMed gnomAD |
|
| rs765397426 | 786 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1186638304 | 787 | A>S | No | gnomAD | |
| rs1186638304 | 787 | A>T | No | gnomAD | |
| TCGA novel | 789 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1231608 rs2033233054 |
790 | L>V | large_intestine [Cosmic] | No |
cosmic curated Ensembl |
| rs1372873747 | 791 | P>T | No |
TOPMed gnomAD |
|
| rs762272441 | 793 | N>H | No |
ExAC TOPMed gnomAD |
|
| rs1259541929 | 793 | N>T | No | gnomAD | |
| TCGA novel | 795 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1419473393 | 796 | N>H | No |
TOPMed gnomAD |
|
| rs2033197111 | 796 | N>T | No | TOPMed | |
| rs2033197079 | 799 | G>V | No | Ensembl | |
| rs141762554 | 801 | E>D | No |
ESP ExAC |
|
|
RCV001955320 rs374815480 |
802 | N>S | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2033197000 | 802 | N>Y | No | Ensembl | |
| rs1567130321 | 804 | I>V | No | Ensembl | |
| TCGA novel | 805 | N>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756130552 | 805 | N>K | No |
ExAC gnomAD |
|
| rs752569032 | 806 | K>E | No |
ExAC gnomAD |
|
| rs750555692 | 815 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs778954996 | 816 | S>* | No |
ExAC gnomAD |
|
| rs778954996 | 816 | S>L | No |
ExAC gnomAD |
|
| rs1050248132 | 817 | G>S | No | TOPMed | |
| rs866504949 | 819 | G>E | No | Ensembl | |
| rs547441773 | 820 | V>I | No | 1000Genomes | |
| rs1286553773 | 822 | E>D | No | gnomAD | |
| rs1452416487 | 823 | L>V | No | gnomAD | |
| rs2033181365 | 825 | D>N | No | TOPMed | |
| rs1160352539 | 827 | S>L | No |
TOPMed gnomAD |
|
| rs757567255 | 829 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
|
COSM961871 rs2033181155 |
830 | D>Y | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs2033181109 | 831 | F>* | No | Ensembl | |
| rs980100953 | 831 | F>V | No | Ensembl | |
| rs764320343 | 832 | N>D | No |
ExAC gnomAD |
|
| rs866636571 | 833 | M>I | No |
1000Genomes gnomAD |
|
| rs2033180924 | 833 | M>T | No | TOPMed | |
| rs760971096 | 833 | M>V | No |
ExAC gnomAD |
|
| rs2033180785 | 837 | H>R | No |
TOPMed gnomAD |
|
| rs2033180816 | 837 | H>Y | No | TOPMed | |
| rs1249484505 | 839 | S>P | No | TOPMed | |
| rs1436164639 | 842 | Q>R | No | gnomAD | |
| rs1485579827 | 843 | H>R | No | gnomAD | |
| rs2033180597 | 844 | S>N | No | Ensembl | |
| COSM3501342 | 845 | K>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1168558964 | 847 | E>K | No | gnomAD | |
| rs757299423 | 848 | H>R | No |
ExAC gnomAD |
|
| rs1195972068 | 849 | M>L | No |
TOPMed gnomAD |
|
| rs1375695034 | 849 | M>T | No | TOPMed | |
| rs1195972068 | 849 | M>V | No |
TOPMed gnomAD |
|
| TCGA novel | 850 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1257419086 | 851 | K>N | No | gnomAD | |
| rs777887212 | 854 | R>K | No |
ExAC gnomAD |
|
| rs756328358 | 856 | Q>K | No |
ExAC gnomAD |
|
| COSM2153434 | 857 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753161247 | 857 | E>K | No |
ExAC gnomAD |
|
|
COSM470662 rs767980027 |
858 | N>K | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated ExAC gnomAD NCI-TCGA Cosmic |
| rs111240413 | 859 | K>T | No | Ensembl | |
| rs2141305888 | 860 | D>H | No | Ensembl | |
| rs756461131 | 862 | A>V | No |
ExAC gnomAD |
|
| rs1339644826 | 863 | L>F | No |
TOPMed gnomAD |
|
| rs781493412 | 864 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs781493412 | 864 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs2033160723 | 864 | P>S | No | TOPMed | |
| rs2033160637 | 865 | P>L | No | TOPMed | |
| rs2033160553 | 866 | P>S | No | gnomAD | |
| rs561769201 | 867 | P>S | No |
1000Genomes TOPMed gnomAD |
|
| rs758919702 | 868 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs766823644 | 868 | P>S | No |
ExAC gnomAD |
|
| rs1181797056 | 870 | E>K | No | gnomAD | |
| rs749961748 | 872 | C>S | No |
ExAC gnomAD |
|
| rs1411186375 | 873 | P>A | No | TOPMed | |
| rs1016115192 | 874 | A>V | No | Ensembl | |
| rs764803016 | 875 | F>L | No |
ExAC gnomAD |
|
|
rs1000752634 RCV002020083 |
877 | K>E | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1000752634 | 877 | K>Q | No |
TOPMed gnomAD |
|
| rs1351071443 | 879 | I>V | No | TOPMed | |
| rs1348382128 | 880 | N>S | No | gnomAD | |
| rs1417283661 | 881 | L>V | No |
TOPMed gnomAD |
|
| rs763689059 | 883 | N>S | No |
ExAC gnomAD |
|
| rs1301011110 | 886 | I>T | No |
TOPMed gnomAD |
|
| rs775322814 | 887 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs2033159487 | 887 | M>T | No | TOPMed | |
| rs1324225323 | 888 | M>I | No |
TOPMed gnomAD |
|
| rs542518072 | 890 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2033159327 | 893 | T>A | No |
TOPMed gnomAD |
|
|
RCV001820735 RCV003718441 rs199513326 |
893 | T>I | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs2033159327 | 893 | T>P | No |
TOPMed gnomAD |
|
| rs141507311 | 894 | V>A | No |
ESP TOPMed gnomAD |
|
| rs375804838 | 894 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs769994336 | 896 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs748177653 | 896 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs2033159056 | 897 | R>Q | No |
TOPMed gnomAD |
|
|
COSM1493442 rs781583486 |
897 | R>W | kidney [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs2033158919 | 899 | I>T | No | Ensembl | |
| rs780456002 | 900 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs546372560 | 900 | D>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 900 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2033158797 | 901 | T>A | No | TOPMed | |
| rs996967856 | 901 | T>I | No | TOPMed | |
| rs898785197 | 903 | S>A | No |
TOPMed gnomAD |
|
| rs758898370 | 904 | N>D | No |
ExAC gnomAD |
|
| rs750948370 | 905 | L>W | No |
ExAC gnomAD |
|
| TCGA novel | 906 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2033158652 | 906 | W>G | No | Ensembl | |
| rs765675717 | 907 | T>A | No | ExAC | |
|
rs2033158510 COSM961870 |
908 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs370452802 | 909 | G>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs2054417593 | 909 | G>R | No | TOPMed | |
| rs763778855 | 910 | M>V | No |
ExAC gnomAD |
|
| rs1438922220 | 913 | M>V | No |
TOPMed gnomAD |
|
| rs746170520 | 914 | A>V | No |
ExAC gnomAD |
|
| rs1342513053 | 917 | I>M | No | gnomAD | |
| rs1414768981 | 926 | K>E | No | gnomAD | |
| rs1360062702 | 927 | Q>* | No |
TOPMed gnomAD |
|
| rs1360062702 | 927 | Q>E | No |
TOPMed gnomAD |
|
| rs2033126996 | 929 | L>F | No |
TOPMed gnomAD |
|
| rs113857535 | 929 | L>P | No | gnomAD | |
| rs899677587 | 931 | K>Q | No | TOPMed | |
| rs757731505 | 931 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs753392602 | 932 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs777250042 | 933 | P>A | No |
ExAC gnomAD |
|
| rs1596106053 | 934 | E>K | No | TOPMed | |
|
COSM700542 rs1415080831 |
938 | T>A | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs755788395 | 938 | T>I | No |
ExAC gnomAD |
|
| rs2141303554 | 940 | D>N | No | Ensembl | |
| rs149061186 | 941 | F>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs767015627 | 943 | H>R | No |
ExAC gnomAD |
|
| COSM3501340 | 943 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033126243 | 944 | K>E | No |
TOPMed gnomAD |
|
| rs759393697 | 945 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1283304268 | 945 | A>V | No | gnomAD | |
| rs2033126120 | 947 | R>* | No | TOPMed | |
| rs2033126120 | 947 | R>G | No | TOPMed | |
| rs767831257 | 947 | R>S | No |
ExAC gnomAD |
|
| rs915536049 | 948 | L>M | No |
TOPMed gnomAD |
|
| rs989734306 | 948 | L>S | No | Ensembl | |
| rs759738235 | 949 | G>R | No |
ExAC gnomAD |
|
| rs929403892 | 950 | S>R | No |
TOPMed gnomAD |
|
| rs774761748 | 951 | S>L | No |
ExAC gnomAD |
|
| rs1292376078 | 952 | A>T | No | gnomAD | |
|
rs959859810 COSM270496 |
952 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs773730651 | 953 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs749593414 | 953 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs926829623 | 953 | M>V | No |
TOPMed gnomAD |
|
| rs1411334425 | 954 | N>S | No | TOPMed | |
| rs770159784 | 955 | I>K | No |
ExAC gnomAD |
|
| rs1400095500 | 956 | Q>* | No |
TOPMed gnomAD |
|
| rs1400095500 | 956 | Q>E | No |
TOPMed gnomAD |
|
| rs1380638654 | 957 | M>I | No | gnomAD | |
| rs2033108218 | 960 | E>K | No | Ensembl | |
| rs755013298 | 962 | L>H | No | Ensembl | |
| rs747733828 | 963 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs747733828 | 963 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs780543525 | 964 | G>R | No |
ExAC gnomAD |
|
| rs145788329 | 969 | E>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs145788329 | 969 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs938503004 | 970 | G>V | No |
TOPMed gnomAD |
|
| rs1287101894 | 975 | I>K | No |
TOPMed gnomAD |
|
| rs1287101894 | 975 | I>T | No |
TOPMed gnomAD |
|
| rs750207413 | 976 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs752882409 | 977 | W>* | No |
ExAC gnomAD |
|
| rs28678764 | 977 | W>G | No | Ensembl | |
| rs767487834 | 979 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs759541918 | 979 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs767487834 | 979 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs2033107296 | 980 | Q>* | No | TOPMed | |
| rs2033107296 | 980 | Q>K | No | TOPMed | |
| rs2033037243 | 981 | M>T | No | TOPMed | |
| rs1382433807 | 983 | D>H | No |
TOPMed gnomAD |
|
| rs777027545 | 984 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs769246548 | 985 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs769246548 | 985 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs112041571 | 986 | K>R | No | Ensembl | |
| rs1249254589 | 987 | R>* | No | gnomAD | |
| rs760305347 | 987 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2033036808 | 989 | R>G | No | Ensembl | |
| rs2033036772 | 991 | K>R | No |
TOPMed gnomAD |
|
| rs775121179 | 993 | C>R | No |
ExAC gnomAD |
|
| rs771910755 | 994 | L>F | No | ExAC | |
| rs778559456 | 996 | V>I | No |
ExAC gnomAD |
|
| rs778559456 | 996 | V>L | No |
ExAC gnomAD |
|
| rs770642267 | 997 | A>V | No |
ExAC gnomAD |
|
| rs1339813285 | 998 | T>N | No | gnomAD | |
| rs2141298487 | 999 | T>I | No | Ensembl | |
| rs756014691 | 1002 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1465347996 | 1004 | S>P | No | gnomAD | |
| rs2033036116 | 1005 | I>V | No | Ensembl | |
| rs148993210 | 1007 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs376036774 | 1007 | N>S | No |
ESP TOPMed gnomAD |
|
| rs1456787822 | 1008 | D>V | No | gnomAD | |
| rs777521212 | 1011 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1239608034 | 1012 | H>R | No | gnomAD | |
| rs2033016441 | 1012 | H>Y | No | gnomAD | |
| TCGA novel | 1013 | D>E | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs79459762 | 1014 | K>N | No | TOPMed | |
| COSM961869 | 1015 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1450509467 | 1015 | E>D | No | gnomAD | |
| rs769779282 | 1015 | E>Q | No |
ExAC gnomAD |
|
| rs748070576 | 1017 | A>S | No |
ExAC gnomAD |
|
| rs748070576 | 1017 | A>T | No |
ExAC gnomAD |
|
| rs267604210 | 1019 | R>G | No |
TOPMed gnomAD |
|
| rs1487286460 | 1019 | R>Q | No |
TOPMed gnomAD |
|
| rs200354286 | 1021 | R>G | No | 1000Genomes | |
| rs2033015587 | 1023 | A>P | No | TOPMed | |
| rs922768419 | 1025 | A>S | No | Ensembl | |
| rs758431812 | 1025 | A>V | No |
ExAC gnomAD |
|
| rs2033015412 | 1026 | A>P | No | gnomAD | |
| rs779045429 | 1027 | R>S | No |
ExAC gnomAD |
|
| rs754217712 | 1029 | H>Y | No |
ExAC TOPMed gnomAD |
|
|
rs1330542956 COSM1708065 |
1030 | R>C | skin [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs764552375 | 1030 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs181657162 | 1032 | K>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM4054764 | 1032 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs966854415 | 1034 | M>L | No | TOPMed | |
| rs966854415 | 1034 | M>V | No | TOPMed | |
| rs2033014908 | 1036 | Q>H | No | TOPMed | |
| rs1322385972 | 1036 | Q>R | No |
TOPMed gnomAD |
|
| rs1022801695 | 1037 | M>I | No | Ensembl | |
| rs1298185944 | 1039 | A>T | No | gnomAD | |
| rs1478286465 | 1043 | N>I | No |
TOPMed gnomAD |
|
| TCGA novel | 1044 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1010932062 | 1045 | I>M | No | Ensembl | |
| rs545826909 | 1045 | I>T | No |
1000Genomes TOPMed gnomAD |
|
| rs1477505340 | 1047 | T>I | No | gnomAD | |
| rs758995090 | 1048 | H>Y | No |
ExAC gnomAD |
|
| rs1200377569 | 1049 | K>I | No |
TOPMed gnomAD |
|
| TCGA novel | 1050 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs373336900 | 1051 | M>L | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs1596102688 | 1051 | M>T | No | Ensembl | |
| rs373336900 | 1051 | M>V | No |
1000Genomes ESP TOPMed gnomAD |
|
| COSM6142139 | 1052 | Y>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1252787304 | 1053 | D>E | No |
TOPMed gnomAD |
|
| rs1262380829 | 1053 | D>H | No | gnomAD | |
| rs2033013754 | 1055 | T>I | No | TOPMed | |
| COSM1301138 | 1056 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033013541 | 1057 | E>D | No | TOPMed | |
| rs1276268836 | 1058 | M>I | No | gnomAD | |
| rs147741987 | 1058 | M>T | No |
ESP ExAC gnomAD |
|
| rs1198901772 | 1060 | G>R | No |
TOPMed gnomAD |
|
| rs773055777 | 1061 | K>R | No |
ExAC gnomAD |
|
| COSM961868 | 1062 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1235503054 | 1064 | S>F | No |
TOPMed gnomAD |
|
| rs1235503054 | 1064 | S>Y | No |
TOPMed gnomAD |
|
| rs776583103 | 1066 | M>L | No |
ExAC TOPMed gnomAD |
|
|
RCV001819325 rs776583103 |
1066 | M>V | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1278091838 | 1067 | E>D | No | gnomAD | |
| rs984068276 | 1068 | E>K | No | TOPMed | |
| rs1156621033 | 1071 | T>N | No | gnomAD | |
| rs770105611 | 1072 | P>A | No |
ExAC gnomAD |
|
| rs1476974247 | 1072 | P>L | No | TOPMed | |
| rs781606224 | 1073 | A>E | No |
ExAC gnomAD |
|
| rs755502795 | 1074 | V>D | No |
ExAC TOPMed gnomAD |
|
| rs2032847491 | 1074 | V>I | No | TOPMed | |
| rs2032847395 | 1075 | S>I | No | gnomAD | |
| rs141543407 | 1075 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs757935803 | 1077 | Y>H | No |
ExAC gnomAD |
|
| rs749905823 | 1078 | S>A | No |
ExAC TOPMed |
|
| rs2141289553 | 1078 | S>C | No | Ensembl | |
| COSM470660 | 1080 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1341572923 | 1081 | A>V | No | gnomAD | |
| rs905594006 | 1084 | P>H | No | TOPMed | |
| rs1294326600 | 1085 | K>Q | No | gnomAD | |
| rs774062223 | 1086 | R>Q | No |
TOPMed gnomAD |
|
| rs764919801 | 1086 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2032846378 | 1087 | G>D | No | Ensembl | |
| rs761442343 | 1089 | S>P | No |
ExAC gnomAD |
|
| rs2032846131 | 1090 | V>I | No | TOPMed | |
| rs753704364 | 1091 | T>I | No |
ExAC gnomAD |
|
| rs200690084 | 1094 | E>A | No |
1000Genomes ExAC gnomAD |
|
| rs775523272 | 1095 | V>G | No |
ExAC gnomAD |
|
| rs1044494026 | 1095 | V>M | No |
TOPMed gnomAD |
|
| rs895447796 | 1098 | C>Y | No | Ensembl | |
| rs773400142 | 1101 | C>S | No | ExAC | |
| rs1275821790 | 1104 | E>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032845163 | 1104 | E>* | No |
TOPMed gnomAD |
|
| rs2032845111 | 1104 | E>G | No | TOPMed | |
|
COSM3987957 rs2032845163 |
1104 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed gnomAD |
| rs2032844943 | 1105 | Q>P | No | TOPMed | |
| rs867111288 | 1107 | V>L | No | Ensembl | |
| rs1357498324 | 1108 | K>E | No | gnomAD | |
| rs368692976 | 1109 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs2032843459 | 1111 | N>S | No |
TOPMed gnomAD |
|
| rs1272008445 | 1112 | N>D | No | TOPMed | |
| rs1159425116 | 1112 | N>K | No | gnomAD | |
| rs781202228 | 1113 | A>T | No |
ExAC gnomAD |
|
| rs2141289474 | 1114 | M>I | No | Ensembl | |
| rs1423068595 | 1114 | M>V | No | gnomAD | |
| rs1190694356 | 1117 | S>* | No |
TOPMed gnomAD |
|
|
rs1190694356 COSM3886999 |
1117 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed gnomAD |
| rs1190694356 | 1117 | S>W | No |
TOPMed gnomAD |
|
|
COSM3501338 rs1271672039 |
1118 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| TCGA novel | 1118 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1215132259 | 1119 | C>F | No |
TOPMed gnomAD |
|
| rs1215132259 | 1119 | C>Y | No |
TOPMed gnomAD |
|
| rs1253300095 | 1120 | V>I | No | TOPMed | |
| rs780652360 | 1122 | K>E | No |
ExAC gnomAD |
|
| rs1032341842 | 1123 | S>F | No | TOPMed | |
| TCGA novel | 1127 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs371256699 | 1128 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs749993781 | 1129 | H>R | No |
ExAC gnomAD |
|
| rs1219292120 | 1131 | G>E | No | gnomAD | |
| rs778393287 | 1132 | K>E | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1132 | K>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs377271561 | 1133 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs377271561 | 1133 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs756693535 | 1133 | P>S | No |
ExAC gnomAD |
|
| rs922034554 | 1134 | I>R | No | gnomAD | |
| rs922034554 | 1134 | I>T | No | gnomAD | |
| rs763608145 | 1134 | I>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1135 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752401371 | 1136 | L>F | No |
ExAC gnomAD |
|
| rs1175699332 | 1137 | S>L | No | gnomAD | |
| rs1431607732 | 1138 | G>R | No | gnomAD | |
| rs1192072036 | 1138 | G>V | No | gnomAD | |
| rs1182077139 | 1139 | E>A | No | gnomAD | |
| rs1182077139 | 1139 | E>G | No | gnomAD | |
| rs777518378 | 1140 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs777518378 | 1140 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2032763873 | 1141 | L>P | No | TOPMed | |
| rs752592051 | 1142 | D>E | No | ExAC | |
| rs2141286265 | 1143 | P>L | No | Ensembl | |
| rs767415187 | 1144 | L>F | No |
ExAC gnomAD |
|
| rs374595653 | 1146 | M>K | No |
ESP TOPMed gnomAD |
|
|
RCV001999493 rs754753489 |
1146 | M>L | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs754753489 | 1146 | M>V | No |
ExAC gnomAD |
|
| rs994453837 | 1149 | D>H | No | gnomAD | |
| rs766747277 | 1149 | D>V | No | Ensembl | |
| rs144681428 | 1150 | L>F | No |
ESP TOPMed |
|
| rs2032763333 | 1154 | T>A | No | Ensembl | |
| rs2032763286 | 1155 | Y>F | No | TOPMed | |
|
rs1422405222 COSM1372963 |
1156 | T>I | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs761994943 | 1158 | S>N | No |
ExAC gnomAD |
|
| rs2032763097 | 1160 | G>V | No | TOPMed | |
| TCGA novel | 1161 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs912375537 | 1163 | M>T | No |
TOPMed gnomAD |
|
|
rs34803922 RCV001193918 |
1164 | H>Q | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs368770602 | 1165 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs368770602 | 1165 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
|
rs772442007 COSM961867 |
1166 | V>M | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
|
TCGA novel rs2032762649 |
1168 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| TCGA novel | 1168 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032762595 | 1169 | Q>H | No | Ensembl | |
| rs1428221545 | 1176 | Q>E | No |
TOPMed gnomAD |
|
| rs1428221545 | 1176 | Q>K | No |
TOPMed gnomAD |
|
| rs373216782 | 1176 | Q>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs373216782 | 1176 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs373216782 | 1176 | Q>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs149935389 | 1177 | L>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs760753358 | 1178 | S>N | No |
ExAC gnomAD |
|
| rs1021094121 | 1178 | S>R | No | Ensembl | |
| rs775714699 | 1179 | S>A | No |
ExAC gnomAD |
|
| rs752737853 | 1179 | S>F | No | Ensembl | |
|
TCGA novel rs2032744227 |
1180 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1169717202 | 1181 | Q>R | No | gnomAD | |
| rs2032744125 | 1182 | R>C | No | TOPMed | |
|
COSM961866 rs139161796 |
1182 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP NCI-TCGA TOPMed gnomAD |
|
rs767690691 COSM3794177 |
1184 | H>Y | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1369978716 | 1186 | D>E | No | gnomAD | |
| rs2032743910 | 1188 | F>V | No |
TOPMed gnomAD |
|
| rs759938108 | 1189 | D>G | No |
ExAC gnomAD |
|
| rs1426906183 | 1190 | L>F | No |
TOPMed gnomAD |
|
| rs1402926607 | 1191 | E>G | No |
TOPMed gnomAD |
|
| rs1359078808 | 1191 | E>K | No | gnomAD | |
| rs1157621442 | 1192 | S>G | No | gnomAD | |
| rs1470328021 | 1192 | S>N | No | gnomAD | |
| rs771521806 | 1194 | E>D | No |
ExAC gnomAD |
|
| rs2032743431 | 1194 | E>DYSLFSVQLHHV* | No | Ensembl | |
| COSM5705874 | 1196 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1346463706 | 1196 | L>V | No |
TOPMed gnomAD |
|
| rs2032743225 | 1198 | P>R | No | TOPMed | |
| rs2032743175 | 1199 | L>H | No | Ensembl | |
| rs1268583387 | 1200 | C>R | No |
TOPMed gnomAD |
|
| rs1269513499 | 1201 | K>R | No | gnomAD | |
| rs773701351 | 1206 | T>I | No |
ExAC gnomAD |
|
| TCGA novel | 1207 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032742761 | 1212 | P>L | No | Ensembl | |
| TCGA novel | 1212 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032742710 | 1213 | L>S | No | TOPMed | |
| rs1284454148 | 1215 | P>L | No | gnomAD | |
| rs769199188 | 1215 | P>S | No |
ExAC gnomAD |
|
| rs1215201109 | 1216 | Q>E | No | TOPMed | |
| rs747644799 | 1217 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs1338845115 | 1218 | I>V | No | gnomAD | |
| TCGA novel | 1219 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1045640755 | 1219 | N>Y | No |
TOPMed gnomAD |
|
| rs1567119831 | 1224 | D>N | No | Ensembl | |
|
rs117347582 RCV001519222 |
1225 | A>S | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs766654821 | 1225 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs760997475 | 1226 | L>H | No | Ensembl | |
| rs2032669870 | 1226 | L>V | No |
TOPMed gnomAD |
|
| rs2032669592 | 1233 | A>T | No | Ensembl | |
| rs776268861 | 1234 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs776268861 | 1234 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
RCV002008526 rs770295197 |
1234 | R>W | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
RCV002017436 rs2032669345 |
1236 | I>M | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1422824446 | 1236 | I>T | No | gnomAD | |
| rs375380714 | 1237 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs746666900 | 1241 | A>V | No |
ExAC gnomAD |
|
| rs187899168 | 1244 | S>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1392640703 | 1245 | G>D | No | gnomAD | |
| rs771788893 | 1246 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs2032668919 | 1247 | N>D | No | TOPMed | |
|
RCV000882861 rs148375509 |
1247 | N>K | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1205633763 | 1250 | H>L | No | gnomAD | |
| rs1205633763 | 1250 | H>R | No | gnomAD | |
| rs1567117427 | 1253 | G>E | No |
TOPMed gnomAD |
|
| rs370903647 | 1254 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1257 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs958032449 | 1258 | P>R | No | Ensembl | |
| rs771728220 | 1258 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1360665697 | 1259 | I>V | No |
TOPMed gnomAD |
|
|
rs745702420 COSM961865 |
1260 | F>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
RCV000971624 rs139408969 |
1264 | G>E | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1379118920 | 1264 | G>R | No |
TOPMed gnomAD |
|
| rs2032531682 | 1265 | M>I | No | Ensembl | |
| rs770917711 | 1265 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs201899517 | 1266 | G>E | No |
1000Genomes ExAC gnomAD |
|
| rs1322889701 | 1267 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1425033820 | 1268 | S>C | No | gnomAD | |
| rs1165655070 | 1273 | H>D | No | gnomAD | |
| rs1042695336 | 1274 | S>F | No |
TOPMed gnomAD |
|
| rs1042695336 | 1274 | S>Y | No |
TOPMed gnomAD |
|
| rs199934267 | 1275 | I>V | No |
TOPMed gnomAD |
|
| rs777627331 | 1276 | L>Q | No |
ExAC gnomAD |
|
| rs1268536076 | 1277 | S>N | No | gnomAD | |
| rs1177836235 | 1278 | F>Y | No | gnomAD | |
| rs774045909 | 1280 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2032530976 | 1281 | E>G | No | TOPMed | |
| COSM433887 | 1281 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs561720791 | 1283 | S>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 1284 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032516500 | 1285 | K>R | No | TOPMed | |
| TCGA novel | 1285 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs749031006 | 1286 | Y>C | No |
ExAC gnomAD |
|
| rs2032516363 | 1288 | N>H | No | TOPMed | |
| rs564247521 | 1289 | S>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM1478130 | 1290 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs149097306 COSM3420308 |
1291 | K>N | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs1217564561 | 1292 | E>D | No |
TOPMed gnomAD |
|
| rs2032516078 | 1292 | E>K | No | Ensembl | |
| rs1303835548 | 1293 | M>I | No | TOPMed | |
| rs1309888623 | 1295 | I>F | No | gnomAD | |
| rs148011271 | 1296 | L>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2032515684 | 1296 | L>P | No | TOPMed | |
| rs1052813763 | 1297 | F>S | No | TOPMed | |
| rs955233917 | 1298 | A>T | No | TOPMed | |
| rs2032515532 | 1298 | A>V | No | TOPMed | |
| rs746117653 | 1300 | T>A | No |
ExAC gnomAD |
|
| rs2032515198 | 1302 | Y>C | No |
TOPMed gnomAD |
|
| TCGA novel | 1303 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM470659 | 1305 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1301137 | 1306 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2032514969 | 1307 | K>Q | No | Ensembl | |
| rs181623854 | 1308 | V>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs149186253 | 1310 | P>R | No |
ESP ExAC gnomAD |
|
| rs1433193595 | 1310 | P>S | No | gnomAD | |
| rs1433193595 | 1310 | P>T | No | gnomAD | |
| rs1269197116 | 1312 | E>K | No |
TOPMed gnomAD |
|
| rs1269197116 | 1312 | E>Q | No |
TOPMed gnomAD |
|
| rs2141276024 | 1313 | R>T | No | Ensembl | |
| rs2032514441 | 1314 | D>E | No | Ensembl | |
| rs778476475 | 1314 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs778476475 | 1314 | D>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1314 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1432080674 | 1315 | P>L | No | gnomAD | |
| TCGA novel | 1315 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs373699629 | 1316 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs373699629 | 1316 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs138510410 | 1316 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV001903318 rs138510410 |
1316 | R>Q | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs767055800 | 1317 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs767055800 | 1317 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs552698480 | 1318 | P>S | No |
TOPMed gnomAD |
|
| rs552698480 | 1318 | P>T | No |
TOPMed gnomAD |
|
| rs751244971 | 1319 | M>I | No |
ExAC gnomAD |
|
| rs1241927414 | 1319 | M>T | No | gnomAD | |
| rs758908861 | 1319 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1462297718 | 1320 | L>P | No |
TOPMed gnomAD |
|
| rs147112447 | 1322 | W>C | No |
ESP TOPMed gnomAD |
|
| rs778270634 | 1326 | A>T | No |
TOPMed gnomAD |
|
| rs541843760 | 1329 | I>L | No |
1000Genomes ExAC gnomAD |
|
| rs2032513624 | 1329 | I>S | No | TOPMed | |
| rs1297229705 | 1331 | A>G | No |
TOPMed gnomAD |
|
|
RCV002040097 rs761815089 |
1332 | I>T | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs1336396878 | 1332 | I>V | No | gnomAD | |
| rs1292268909 | 1336 | L>S | No |
TOPMed gnomAD |
|
| rs1292268909 | 1336 | L>W | No |
TOPMed gnomAD |
|
|
rs1340343438 COSM961864 |
1337 | G>E | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1311071387 | 1337 | G>R | No |
TOPMed gnomAD |
|
| rs372661227 | 1338 | D>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1001855717 | 1340 | G>R | No | gnomAD | |
| rs866017422 | 1342 | P>L | No |
TOPMed gnomAD |
|
| rs2032435762 | 1344 | F>L | No | TOPMed | |
| rs141869423 | 1348 | Q>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs781741948 | 1349 | N>K | No |
ExAC gnomAD |
|
| rs1468892016 | 1352 | H>R | No | gnomAD | |
| rs1767559409 | 1353 | N>S | No | gnomAD | |
| rs757935824 | 1357 | A>P | No |
ExAC gnomAD |
|
| rs2032422322 | 1357 | A>V | No | Ensembl | |
|
rs143022130 RCV000901283 |
1359 | M>I | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs745352887 | 1359 | M>V | No |
ExAC gnomAD |
|
| rs1464967921 | 1361 | F>L | No |
TOPMed gnomAD |
|
| rs1464967921 | 1361 | F>V | No |
TOPMed gnomAD |
|
| rs2032421978 | 1362 | A>T | No | Ensembl | |
| rs900840781 | 1363 | V>G | No | Ensembl | |
| rs2032421933 | 1363 | V>L | No | Ensembl | |
|
COSM188027 rs557063833 |
1364 | A>T | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC gnomAD |
| rs1042166926 | 1365 | Q>* | No |
TOPMed gnomAD |
|
| rs1567115645 | 1365 | Q>R | No | Ensembl | |
| rs2032421706 | 1366 | R>G | No | TOPMed | |
| rs1169079077 | 1366 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs753736868 | 1368 | T>I | No |
ExAC gnomAD |
|
| rs763804672 | 1369 | C>R | No |
ExAC gnomAD |
|
| rs755910310 | 1370 | P>R | No |
ExAC gnomAD |
|
|
COSM344351 rs752699712 |
1371 | Q>E | lung [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1221457511 | 1371 | Q>R | No |
TOPMed gnomAD |
|
| COSM1372962 | 1373 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2032421232 | 1373 | L>V | No | TOPMed | |
| rs1227628815 | 1374 | I>K | No |
TOPMed gnomAD |
|
| rs1269984047 | 1374 | I>L | No |
TOPMed gnomAD |
|
| rs1227628815 | 1374 | I>T | No |
TOPMed gnomAD |
|
| rs1269984047 | 1374 | I>V | No |
TOPMed gnomAD |
|
| rs1327821931 | 1375 | Q>R | No |
TOPMed gnomAD |
|
| rs763162821 | 1377 | H>Y | No |
ExAC gnomAD |
|
| rs773402048 | 1378 | L>V | No |
ExAC TOPMed gnomAD |
|
|
rs372200839 RCV000890155 |
1380 | R>C | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs536630830 COSM1129202 |
1380 | R>H | prostate [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs372200839 | 1380 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1297366018 | 1381 | L>P | No |
TOPMed gnomAD |
|
| rs16957294 | 1382 | L>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2032315753 | 1386 | L>F | No | Ensembl | |
| rs2032315714 | 1387 | P>R | No | gnomAD | |
| rs754951007 | 1388 | N>K | No |
ExAC gnomAD |
|
|
RCV000927613 rs546603068 |
1389 | I>L | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs761894888 | 1389 | I>M | No |
ExAC gnomAD |
|
| rs765236578 | 1389 | I>R | No |
ExAC TOPMed gnomAD |
|
| rs546603068 | 1389 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs7175979 | 1392 | E>Q | No | Ensembl | |
| rs754037799 | 1393 | D>H | No |
ExAC gnomAD |
|
| rs1321733993 | 1394 | T>I | No |
TOPMed gnomAD |
|
| rs2032315204 | 1395 | P>Q | No | Ensembl | |
| rs2032315156 | 1396 | C>Y | No | TOPMed | |
| rs764268195 | 1397 | L>H | No |
ExAC gnomAD |
|
| rs761074877 | 1400 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1232782722 | 1402 | L>V | No | gnomAD | |
| rs2032314899 | 1403 | F>L | No | TOPMed | |
| rs2032314855 | 1404 | H>L | No | TOPMed | |
| rs1391489277 | 1406 | L>W | No |
TOPMed gnomAD |
|
| rs2032298424 | 1407 | V>G | No | gnomAD | |
| rs2032298478 | 1407 | V>M | No | Ensembl | |
|
TCGA novel rs2032298374 |
1408 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs757320774 | 1409 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs368739715 | 1410 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs952328172 | 1410 | V>M | No | TOPMed | |
| rs764355906 | 1412 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs1321319018 | 1412 | A>T | No | gnomAD | |
| rs764355906 | 1412 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs760880459 | 1413 | F>L | No |
ExAC gnomAD |
|
| rs2032297827 | 1414 | P>A | No | gnomAD | |
| rs2032297827 | 1414 | P>S | No | gnomAD | |
| COSM3501337 | 1415 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1295460185 | 1416 | L>F | No | gnomAD | |
| rs1248081326 | 1417 | Y>C | No |
TOPMed gnomAD |
|
| rs1208073289 | 1417 | Y>H | No |
TOPMed gnomAD |
|
| COSM4397136 | 1418 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs149778120 | 1418 | W>R | No |
ESP ExAC gnomAD |
|
| rs759891752 | 1419 | D>H | No |
ExAC TOPMed gnomAD |
|
|
COSM3401739 rs1596082433 |
1419 | D>V | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| COSM3816107 | 1420 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM3816106 | 1420 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs771441856 | 1422 | V>A | No |
ExAC gnomAD |
|
| rs762422694 | 1425 | Q>* | No |
ExAC TOPMed gnomAD |
|
| rs762422694 | 1425 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs769639574 | 1428 | S>T | No |
ExAC gnomAD |
|
| rs1485349055 | 1430 | S>T | No | gnomAD | |
| rs35242751 | 1431 | S>F | No | Ensembl | |
| rs2032296686 | 1432 | S>F | No | gnomAD | |
| rs768594604 | 1434 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs779903626 | 1435 | H>R | No |
ExAC gnomAD |
|
| rs1313701761 | 1437 | Y>C | No | gnomAD | |
| rs1313701761 | 1437 | Y>F | No | gnomAD | |
| rs200770305 | 1440 | H>R | No | Ensembl | |
| rs758341705 | 1440 | H>Y | No |
ExAC gnomAD |
|
| rs1361518761 | 1443 | T>S | No | gnomAD | |
| rs2032295918 | 1445 | A>T | No | gnomAD | |
| rs1567113488 | 1447 | M>T | No | Ensembl | |
| rs370164043 | 1448 | L>F | No |
ESP TOPMed gnomAD |
|
|
rs777831990 COSM1188987 |
1449 | Q>E | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs151057109 | 1450 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs756125307 | 1450 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1404029287 | 1450 | I>V | No | gnomAD | |
| rs2032295498 | 1451 | L>V | No |
TOPMed gnomAD |
|
| rs1302419167 | 1452 | L>F | No | TOPMed | |
| rs1380142442 | 1453 | T>K | No | TOPMed | |
| rs2141266911 | 1453 | T>S | No | Ensembl | |
| rs755342723 | 1454 | V>A | No |
ExAC gnomAD |
|
| rs1168761400 | 1454 | V>I | No |
TOPMed gnomAD |
|
| rs1168761400 | 1454 | V>L | No |
TOPMed gnomAD |
|
| rs2141266895 | 1455 | D>E | No | Ensembl | |
| rs1390372077 | 1455 | D>N | No | gnomAD | |
| rs1040911348 | 1456 | T>A | No | Ensembl | |
| rs973556447 | 1457 | G>D | No | TOPMed | |
| rs2032188541 | 1460 | L>V | No | TOPMed | |
| rs758633734 | 1462 | Q>H | No |
ExAC gnomAD |
|
| rs112540567 | 1462 | Q>L | No | Ensembl | |
| rs747491054 | 1463 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs765799187 | 1464 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs1244166418 | 1466 | D>A | No |
TOPMed gnomAD |
|
| TCGA novel | 1466 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1323634286 | 1467 | S>G | No |
TOPMed gnomAD |
|
| TCGA novel | 1467 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032188180 | 1467 | S>N | No | TOPMed | |
| rs2141262655 | 1467 | S>R | No | Ensembl | |
| rs1157361706 | 1469 | E>D | No |
TOPMed gnomAD |
|
| rs1446922252 | 1469 | E>Q | No | TOPMed | |
| rs776266897 | 1470 | A>G | No |
ExAC gnomAD |
|
| rs963190749 | 1470 | A>P | No |
TOPMed gnomAD |
|
| rs963190749 | 1470 | A>S | No |
TOPMed gnomAD |
|
| rs2032187921 | 1472 | S>C | No | Ensembl | |
| COSM1756899 | 1472 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs771920019 | 1473 | A>E | No |
ExAC gnomAD |
|
| rs139296068 | 1473 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1470818 rs139296068 |
1473 | A>T | Variant assessed as Somatic; MODERATE impact. prostate [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1364529903 | 1474 | S>T | No | gnomAD | |
| rs780634792 | 1476 | F>C | No |
ExAC TOPMed gnomAD |
|
|
TCGA novel rs2032187406 |
1476 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1277702112 | 1479 | E>G | No | TOPMed | |
| rs2032187125 | 1480 | I>T | No | TOPMed | |
| rs770836072 | 1480 | I>V | No |
ExAC gnomAD |
|
| rs748288799 | 1483 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs781330290 | 1484 | T>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1486 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032104680 | 1487 | S>F | No |
TOPMed gnomAD |
|
| rs747020243 | 1488 | I>N | No |
ExAC gnomAD |
|
| rs747020243 | 1488 | I>T | No |
ExAC gnomAD |
|
| rs772966664 | 1488 | I>V | No |
ExAC gnomAD |
|
| rs780364099 | 1489 | G>E | No |
ExAC gnomAD |
|
| rs2032104477 | 1489 | G>R | No | Ensembl | |
| rs2032104381 | 1490 | C>R | No | Ensembl | |
| rs146245818 | 1492 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1373275225 | 1493 | P>S | No | Ensembl | |
| rs1428742525 | 1494 | G>A | No |
TOPMed gnomAD |
|
| rs1428742525 | 1494 | G>D | No |
TOPMed gnomAD |
|
| rs1596078215 | 1496 | Y>D | No | Ensembl | |
| rs2032104036 | 1498 | W>* | No | gnomAD | |
| rs746301219 | 1504 | G>D | No |
ExAC gnomAD |
|
| rs1460339092 | 1505 | I>V | No | gnomAD | |
| rs1269758301 | 1506 | T>S | No | Ensembl | |
| COSM6077122 | 1507 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757635600 | 1507 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs2032103437 | 1508 | Y>C | No | Ensembl | |
|
rs1017583113 COSM4054763 |
1510 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs778098996 | 1510 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2141259173 | 1511 | C>R | No | Ensembl | |
| rs570338626 | 1512 | A>G | No |
1000Genomes ExAC gnomAD |
|
| rs752129789 | 1513 | A>S | No |
ExAC gnomAD |
|
| rs2032103035 | 1517 | H>Q | No | gnomAD | |
| rs767262512 | 1518 | Y>C | No |
ExAC gnomAD |
|
| rs759187953 | 1519 | L>F | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1519 | L>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1190433724 | 1520 | L>F | No |
TOPMed gnomAD |
|
| rs1273405442 | 1520 | L>P | No |
TOPMed gnomAD |
|
| rs373152125 | 1521 | G>R | No | Ensembl | |
| rs1228725947 | 1522 | V>A | No | gnomAD | |
| TCGA novel | 1522 | V>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs751148703 | 1524 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs762689115 | 1525 | P>L | No |
ExAC gnomAD |
|
| rs2032102428 | 1526 | E>A | No | gnomAD | |
| rs547399909 | 1526 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs953911188 | 1528 | L>R | No |
TOPMed gnomAD |
|
| rs760865862 | 1530 | T>A | No |
ExAC gnomAD |
|
| rs2032102164 | 1530 | T>S | No | TOPMed | |
| rs2032102096 | 1531 | N>D | No |
TOPMed gnomAD |
|
| rs2032102096 | 1531 | N>H | No |
TOPMed gnomAD |
|
| rs765285690 | 1532 | S>F | No |
ExAC gnomAD |
|
|
rs753040283 RCV001997108 |
1534 | E>missing | No |
ClinVar dbSNP |
|
| rs2141257605 | 1534 | E>G | No | Ensembl | |
| rs761660385 | 1534 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs745607297 | 1535 | G>R | No |
TOPMed gnomAD |
|
| rs1567109671 | 1536 | E>D | No | Ensembl | |
| rs1451610441 | 1537 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1330852073 | 1538 | S>N | No | gnomAD | |
| rs201384130 | 1538 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767726031 | 1539 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs143431275 | 1539 | A>V | No |
ESP TOPMed gnomAD |
|
| rs1421736259 | 1540 | L>H | No | gnomAD | |
| rs778825938 | 1540 | L>V | No | Ensembl | |
| rs1025118531 | 1542 | S>N | No | TOPMed | |
| rs1436035164 | 1543 | Y>* | No | gnomAD | |
| rs774750942 | 1543 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs749145487 | 1544 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1012781095 | 1545 | S>F | No |
TOPMed gnomAD |
|
|
RCV001660378 rs3917223 CA7517821 VAR_061822 RCV000249076 |
1548 | T>A | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs3917223 | 1548 | T>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1160622747 | 1549 | N>D | No | gnomAD | |
| rs1160622747 | 1549 | N>H | No | gnomAD | |
| TCGA novel | 1551 | F>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032064219 | 1551 | F>V | No | Ensembl | |
| rs773449503 | 1553 | L>I | No |
ExAC gnomAD |
|
| TCGA novel | 1556 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1212994690 | 1557 | Y>C | No | gnomAD | |
| rs1320146415 | 1557 | Y>H | No |
TOPMed gnomAD |
|
| rs748620426 | 1558 | W>R | No |
ExAC gnomAD |
|
| rs1405769158 | 1559 | D>G | No |
TOPMed gnomAD |
|
| rs1268634399 | 1559 | D>N | No | gnomAD | |
| rs1405769158 | 1559 | D>V | No |
TOPMed gnomAD |
|
| rs2141257505 | 1561 | V>A | No | Ensembl | |
| rs201595040 | 1561 | V>I | No |
1000Genomes ExAC gnomAD |
|
| rs1015374908 | 1563 | P>H | No |
TOPMed gnomAD |
|
| rs1015374908 | 1563 | P>L | No |
TOPMed gnomAD |
|
| rs201922951 | 1565 | L>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs757981667 | 1566 | Q>E | No |
ExAC gnomAD |
|
| rs776252881 | 1566 | Q>R | No | Ensembl | |
| rs1286146322 | 1568 | W>* | No |
TOPMed gnomAD |
|
| rs1567108628 | 1568 | W>C | No | Ensembl | |
| rs1286146322 | 1568 | W>L | No |
TOPMed gnomAD |
|
| rs1567108622 | 1569 | C>Y | No |
TOPMed gnomAD |
|
| rs551157608 | 1570 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1169902581 | 1571 | D>H | No | gnomAD | |
| rs2032000439 | 1572 | P>A | No | Ensembl | |
| TCGA novel | 1572 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1442311785 | 1573 | A>V | No |
TOPMed gnomAD |
|
| rs753658848 | 1574 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs2141255251 | 1575 | L>I | No | Ensembl | |
| rs777911334 | 1577 | C>G | No |
ExAC gnomAD |
|
| rs1484091702 | 1577 | C>Y | No |
TOPMed gnomAD |
|
| rs2032000170 | 1578 | L>V | No | Ensembl | |
| rs2032000106 | 1578 | L>W | No | Ensembl | |
| rs1230988545 | 1579 | K>* | No | gnomAD | |
| rs1472904185 | 1579 | K>R | No |
TOPMed gnomAD |
|
| rs2031999900 | 1580 | Q>* | No | TOPMed | |
| rs1204243548 | 1580 | Q>R | No | gnomAD | |
| rs1277566495 | 1581 | K>* | No | Ensembl | |
| rs145929255 | 1582 | N>H | No |
ESP ExAC TOPMed gnomAD |
|
| COSM2187807 | 1582 | N>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4615557 | 1582 | N>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2031999671 | 1583 | T>A | No |
TOPMed gnomAD |
|
| rs750644112 | 1584 | V>G | No |
ExAC gnomAD |
|
| rs147296247 | 1584 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs147296247 | 1584 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 1585 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765468511 | 1585 | V>G | No |
ExAC gnomAD |
|
| rs765437647 | 1587 | Y>F | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1588 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1389632292 | 1590 | K>N | No |
TOPMed gnomAD |
|
| rs754000018 | 1592 | N>S | No |
ExAC gnomAD |
|
| rs2031952572 | 1593 | S>G | No | Ensembl | |
| rs1459055373 | 1596 | E>A | No | gnomAD | |
| rs2031952465 | 1599 | D>A | No | TOPMed | |
| rs2031952352 | 1600 | D>V | No | Ensembl | |
| rs1596074896 | 1605 | L>R | No | Ensembl | |
| rs1184191714 | 1608 | A>T | No | gnomAD | |
| rs775900069 | 1610 | H>D | No |
ExAC gnomAD |
|
| rs775900069 | 1610 | H>Y | No |
ExAC gnomAD |
|
|
RCV000502460 CA7517761 rs78948790 RCV000953335 |
1612 | R>G | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2031951750 | 1612 | R>K | No | TOPMed | |
| rs757277338 | 1613 | C>F | No |
ExAC gnomAD |
|
| rs754090022 | 1614 | P>A | No |
ExAC gnomAD |
|
| TCGA novel | 1614 | P>H | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754090022 | 1614 | P>S | No |
ExAC gnomAD |
|
|
rs2031850219 TCGA novel |
1615 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1296198050 | 1615 | R>W | No | gnomAD | |
|
RCV001947898 rs764269850 |
1616 | S>F | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs995680412 | 1616 | S>T | No | TOPMed | |
| rs1367261346 | 1617 | A>T | No | gnomAD | |
| rs753088815 | 1619 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs753088815 | 1619 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs767972972 | 1620 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs774666642 | 1621 | R>* | No |
ExAC gnomAD |
|
| rs752483268 | 1621 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs200344888 | 1623 | H>Q | No |
1000Genomes TOPMed gnomAD |
|
|
RCV001911925 rs373539628 |
1623 | H>R | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs773042073 | 1625 | V>I | No |
ExAC gnomAD |
|
| rs1244281789 | 1628 | L>V | No | gnomAD | |
| rs150744789 | 1629 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV001938054 rs141227308 |
1632 | A>V | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs746987173 | 1633 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs768271024 | 1633 | I>V | No |
ExAC gnomAD |
|
| rs779813686 | 1640 | C>W | No |
ExAC gnomAD |
|
| rs2031848696 | 1641 | C>R | No | gnomAD | |
| rs1478610681 | 1642 | Q>E | No |
TOPMed gnomAD |
|
| rs2031848572 | 1644 | I>V | No | TOPMed | |
| rs1228475850 | 1646 | N>T | No | gnomAD | |
| rs1866087835 | 1647 | G>E | No | Ensembl | |
| rs1282285649 | 1647 | G>R | No |
TOPMed gnomAD |
|
| rs756319647 | 1649 | E>D | No |
ExAC TOPMed gnomAD |
|
|
rs1335908378 RCV002026862 |
1649 | E>G | No |
ClinVar dbSNP gnomAD |
|
| rs752793764 | 1650 | V>D | No |
ExAC gnomAD |
|
| rs781632415 | 1653 | C>W | No |
ExAC gnomAD |
|
| rs2031847800 | 1654 | I>V | No | TOPMed | |
| COSM3501335 | 1656 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs550798440 | 1657 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs550798440 COSM4054762 |
1657 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs766965497 | 1657 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1567106185 | 1658 | L>F | No | TOPMed | |
| rs1472745261 | 1659 | H>Q | No | gnomAD | |
| rs1336388891 | 1663 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs749939696 | 1666 | I>V | No |
ExAC gnomAD |
|
| rs113020762 | 1669 | K>N | No |
ExAC TOPMed gnomAD |
|
| rs1407730007 | 1669 | K>Q | No | TOPMed | |
|
rs2141248696 RCV001913822 |
1670 | I>missing | No |
ClinVar dbSNP |
|
| rs1274902485 | 1670 | I>L | No |
TOPMed gnomAD |
|
| rs1274902485 | 1670 | I>V | No |
TOPMed gnomAD |
|
| rs1206778631 | 1674 | R>* | No | gnomAD | |
| rs1303434876 | 1674 | R>Q | No |
TOPMed gnomAD |
|
| rs2031813045 | 1675 | V>A | No | Ensembl | |
| rs758969172 | 1675 | V>M | No |
ExAC gnomAD |
|
| COSM1470819 | 1676 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2031812871 | 1678 | V>I | No |
TOPMed gnomAD |
|
| rs753566038 | 1679 | E>G | No |
ExAC gnomAD |
|
| rs1272137019 | 1679 | E>Q | No |
TOPMed gnomAD |
|
| rs1435775042 | 1682 | A>P | No | gnomAD | |
| rs763632618 | 1682 | A>V | No |
ExAC gnomAD |
|
| rs760576366 | 1683 | R>G | No |
ExAC gnomAD |
|
| rs2031812146 | 1684 | G>S | No | TOPMed | |
| rs1567105631 | 1685 | C>R | No | Ensembl | |
| rs2031811892 | 1686 | A>P | No | gnomAD | |
| rs558013224 | 1687 | Y>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs771784188 | 1687 | Y>D | No |
ExAC TOPMed gnomAD |
|
| rs771784188 | 1687 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs908923221 | 1688 | P>L | No | Ensembl | |
| rs774222169 | 1688 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1016786593 | 1690 | P>S | No | TOPMed | |
| rs1234589430 | 1691 | Y>H | No |
TOPMed gnomAD |
|
| TCGA novel | 1691 | Y>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2031811142 | 1692 | L>V | No | TOPMed | |
| rs2031811068 | 1693 | D>H | No | Ensembl | |
| TCGA novel | 1693 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1192508790 | 1694 | E>A | No |
TOPMed gnomAD |
|
| rs1192508790 | 1694 | E>G | No |
TOPMed gnomAD |
|
| rs1171647817 | 1695 | Y>C | No | gnomAD | |
|
rs748150323 RCV001925729 |
1698 | T>A | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs2031810571 | 1698 | T>K | No | gnomAD | |
| rs1464362805 | 1700 | P>T | No | gnomAD | |
| rs2031810267 | 1701 | G>D | No | gnomAD | |
| rs774315003 | 1704 | R>M | No |
ExAC gnomAD |
|
| rs766208664 | 1705 | G>D | No |
ExAC gnomAD |
|
|
RCV001358080 rs2141245368 |
1705 | G>S | No |
ClinVar Ensembl dbSNP |
|
| rs776569338 | 1706 | N>S | No |
ExAC gnomAD |
|
| rs768716127 | 1707 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs768716127 | 1707 | P>R | No |
ExAC TOPMed gnomAD |
|
| COSM2187798 | 1708 | L>F | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1798392718 | 1709 | H>N | No | TOPMed | |
| COSM5595530 | 1709 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs2031716308 RCV001983107 |
1710 | L>V | No |
ClinVar TOPMed dbSNP |
|
| rs1381831527 | 1711 | S>C | No | gnomAD | |
| rs772603727 | 1712 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs772603727 | 1712 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs141828250 | 1712 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs772603727 | 1712 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs749732238 | 1713 | E>A | No |
ExAC gnomAD |
|
| rs1305811195 | 1714 | R>Q | No |
TOPMed gnomAD |
|
| rs777158316 | 1714 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs771939192 | 1715 | Y>F | No | Ensembl | |
| rs767128461 | 1716 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM1231607 rs201159906 |
1716 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1421959639 | 1717 | K>R | No | gnomAD | |
|
COSM1231606 rs754820204 |
1718 | L>F | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs2031715295 | 1719 | H>R | No | Ensembl | |
| rs1359739147 | 1721 | V>G | No | Ensembl | |
| rs891169475 | 1727 | I>T | No | Ensembl | |
| rs762804188 | 1727 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs2031714973 | 1728 | I>M | No | TOPMed | |
| rs773145191 | 1728 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1482743727 | 1730 | E>A | No | gnomAD | |
| rs764105557 | 1730 | E>Q | No |
ExAC gnomAD |
|
| COSM961862 | 1732 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1183984866 | 1732 | A>V | No |
TOPMed gnomAD |
|
| rs775902455 | 1734 | S>N | No | ExAC | |
|
RCV002082501 rs139367236 |
1734 | S>R | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs774579159 | 1736 | E>K | No |
ExAC gnomAD |
|
| rs749778686 | 1737 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs749778686 | 1737 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs149137306 | 1738 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV001888282 rs2141245282 |
1740 | M>I | No |
ClinVar Ensembl dbSNP |
|
| rs1596070027 | 1740 | M>V | No | Ensembl | |
| rs769337008 | 1741 | L>S | No |
ExAC gnomAD |
|
| rs747638794 | 1742 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1874145198 | 1742 | F>L | No | Ensembl | |
| rs1455989940 | 1745 | N>S | No |
TOPMed gnomAD |
|
| rs1323689207 | 1748 | L>* | No | gnomAD | |
| rs780883781 | 1748 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs201714735 | 1749 | L>V | No | Ensembl | |
| rs2031713820 | 1750 | L>E | No | Ensembl |
1 associated diseases with Q8IWV7
[MIM: 243800]: Johanson-Blizzard syndrome (JBS)
This disorder includes congenital exocrine pancreatic insufficiency, multiple malformations such as nasal wing aplasia, and intellectual disability. Pancreas of individuals with JBS do not express UBR1 and show intrauterine-onset destructive pancreatitis. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- This disorder includes congenital exocrine pancreatic insufficiency, multiple malformations such as nasal wing aplasia, and intellectual disability. Pancreas of individuals with JBS do not express UBR1 and show intrauterine-onset destructive pancreatitis. . Note=The disease is caused by variants affecting the gene represented in this entry.
4 regional properties for Q8IWV7
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.27 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| proteasome complex | A large multisubunit complex which catalyzes protein degradation, found in eukaryotes, archaea and some bacteria. In eukaryotes, this complex consists of the barrel shaped proteasome core complex and one or two associated proteins or complexes that act in regulating entry into or exit from the core. |
| ubiquitin ligase complex | A protein complex that includes a ubiquitin-protein ligase and enables ubiquitin protein ligase activity. The complex also contains other proteins that may confer substrate specificity on the complex. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| leucine binding | Binding to 2-amino-4-methylpentanoic acid. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond |
| zinc ion binding | Binding to a zinc ion (Zn). |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to leucine | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a leucine stimulus. |
| negative regulation of TOR signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of TOR signaling. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| ubiquitin-dependent protein catabolic process via the N-end rule pathway | The chemical reactions and pathways resulting in the breakdown of a protein or peptide covalently tagged with ubiquitin, via the N-end rule pathway. In the N-end rule pathway, destabilizing N-terminal residues (N-degrons) in substrates are recognized by E3 ligases (N-recognins), whereupon the substrates are linked to ubiquitin and then delivered to the proteasome for degradation. |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P19812 | UBR1 | E3 ubiquitin-protein ligase UBR1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | EV |
| Q9VX91 | Ubr1 | E3 ubiquitin-protein ligase UBR1 | Drosophila melanogaster (Fruit fly) | PR |
| Q8IWV8 | UBR2 | E3 ubiquitin-protein ligase UBR2 | Homo sapiens (Human) | SS |
| Q6WKZ8 | Ubr2 | E3 ubiquitin-protein ligase UBR2 | Mus musculus (Mouse) | SS |
| O70481 | Ubr1 | E3 ubiquitin-protein ligase UBR1 | Mus musculus (Mouse) | EV |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MADEEAGGTE | RMEISAELPQ | TPQRLASWWD | QQVDFYTAFL | HHLAQLVPEI | YFAEMDPDLE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KQEESVQMSI | FTPLEWYLFG | EDPDICLEKL | KHSGAFQLCG | RVFKSGETTY | SCRDCAIDPT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| CVLCMDCFQD | SVHKNHRYKM | HTSTGGGFCD | CGDTEAWKTG | PFCVNHEPGR | AGTIKENSRC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PLNEEVIVQA | RKIFPSVIKY | VVEMTIWEEE | KELPPELQIR | EKNERYYCVL | FNDEHHSYDH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VIYSLQRALD | CELAEAQLHT | TAIDKEGRRA | VKAGAYAACQ | EAKEDIKSHS | ENVSQHPLHV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| EVLHSEIMAH | QKFALRLGSW | MNKIMSYSSD | FRQIFCQACL | REEPDSENPC | LISRLMLWDA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| KLYKGARKIL | HELIFSSFFM | EMEYKKLFAM | EFVKYYKQLQ | KEYISDDHDR | SISITALSVQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| MFTVPTLARH | LIEEQNVISV | ITETLLEVLP | EYLDRNNKFN | FQGYSQDKLG | RVYAVICDLK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YILISKPTIW | TERLRMQFLE | GFRSFLKILT | CMQGMEEIRR | QVGQHIEVDP | DWEAAIAIQM |
| 550 | 560 | 570 | 580 | 590 | 600 |
| QLKNILLMFQ | EWCACDEELL | LVAYKECHKA | VMRCSTSFIS | SSKTVVQSCG | HSLETKSYRV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SEDLVSIHLP | LSRTLAGLHV | RLSRLGAVSR | LHEFVSFEDF | QVEVLVEYPL | RCLVLVAQVV |
| 670 | 680 | 690 | 700 | 710 | 720 |
| AEMWRRNGLS | LISQVFYYQD | VKCREEMYDK | DIIMLQIGAS | LMDPNKFLLL | VLQRYELAEA |
| 730 | 740 | 750 | 760 | 770 | 780 |
| FNKTISTKDQ | DLIKQYNTLI | EEMLQVLIYI | VGERYVPGVG | NVTKEEVTMR | EIIHLLCIEP |
| 790 | 800 | 810 | 820 | 830 | 840 |
| MPHSAIAKNL | PENENNETGL | ENVINKVATF | KKPGVSGHGV | YELKDESLKD | FNMYFYHYSK |
| 850 | 860 | 870 | 880 | 890 | 900 |
| TQHSKAEHMQ | KKRRKQENKD | EALPPPPPPE | FCPAFSKVIN | LLNCDIMMYI | LRTVFERAID |
| 910 | 920 | 930 | 940 | 950 | 960 |
| TDSNLWTEGM | LQMAFHILAL | GLLEEKQQLQ | KAPEEEVTFD | FYHKASRLGS | SAMNIQMLLE |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| KLKGIPQLEG | QKDMITWILQ | MFDTVKRLRE | KSCLIVATTS | GSESIKNDEI | THDKEKAERK |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| RKAEAARLHR | QKIMAQMSAL | QKNFIETHKL | MYDNTSEMPG | KEDSIMEEES | TPAVSDYSRI |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| ALGPKRGPSV | TEKEVLTCIL | CQEEQEVKIE | NNAMVLSACV | QKSTALTQHR | GKPIELSGEA |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| LDPLFMDPDL | AYGTYTGSCG | HVMHAVCWQK | YFEAVQLSSQ | QRIHVDLFDL | ESGEYLCPLC |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| KSLCNTVIPI | IPLQPQKINS | ENADALAQLL | TLARWIQTVL | ARISGYNIRH | AKGENPIPIF |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| FNQGMGDSTL | EFHSILSFGV | ESSIKYSNSI | KEMVILFATT | IYRIGLKVPP | DERDPRVPML |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| TWSTCAFTIQ | AIENLLGDEG | KPLFGALQNR | QHNGLKALMQ | FAVAQRITCP | QVLIQKHLVR |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| LLSVVLPNIK | SEDTPCLLSI | DLFHVLVGAV | LAFPSLYWDD | PVDLQPSSVS | SSYNHLYLFH |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| LITMAHMLQI | LLTVDTGLPL | AQVQEDSEEA | HSASSFFAEI | SQYTSGSIGC | DIPGWYLWVS |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| LKNGITPYLR | CAALFFHYLL | GVTPPEELHT | NSAEGEYSAL | CSYLSLPTNL | FLLFQEYWDT |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| VRPLLQRWCA | DPALLNCLKQ | KNTVVRYPRK | RNSLIELPDD | YSCLLNQASH | FRCPRSADDE |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| RKHPVLCLFC | GAILCSQNIC | CQEIVNGEEV | GACIFHALHC | GAGVCIFLKI | RECRVVLVEG |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| KARGCAYPAP | YLDEYGETDP | GLKRGNPLHL | SRERYRKLHL | VWQQHCIIEE | IARSQETNQM |
| LFGFNWQLL |