Q8IUE1
Gene name |
TGIF2LX (TGIFLX) |
Protein name |
Homeobox protein TGIF2LX |
Names |
TGF-beta-induced transcription factor 2-like protein, TGFB-induced factor 2-like protein, X-linked, TGIF-like on the X |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:90316 |
EC number |
|
Protein Class |
|
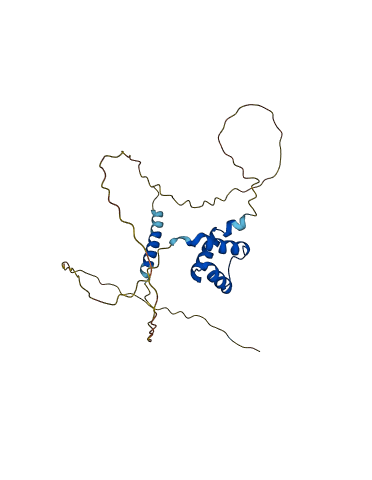
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q8IUE1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2DMN | NMR | - | A | 51-120 | PDB |
| AF-Q8IUE1-F1 | Predicted | AlphaFoldDB |
148 variants for Q8IUE1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA413970332 rs1311902088 |
4 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1569307661 CA413970340 |
5 | A>T | No |
ClinGen Ensembl |
|
|
rs773985382 COSM3694746 CA332384221 |
5 | A>V | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated Ensembl NCI-TCGA |
|
rs866258730 CA413970351 |
7 | G>R | No |
ClinGen gnomAD |
|
|
rs866258730 CA332384225 |
7 | G>S | No |
ClinGen gnomAD |
|
|
rs764080796 CA10466632 |
8 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA10466633 rs764080796 |
8 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs11575111 CA10466634 |
9 | A>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs11575111 CA10466635 |
9 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA413970377 rs1482476387 |
11 | T>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA413970385 rs1349846385 |
12 | Q>H | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 12 | Q>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1000242752 CA332384229 |
13 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
rs756446097 CA10466638 |
14 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs756446097 CA10466637 |
14 | P>Q | No |
ClinGen ExAC gnomAD |
|
|
rs758000770 CA10466640 |
16 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10466641 rs777303673 |
17 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA10466643 rs746477372 |
17 | K>N | No |
ClinGen ExAC |
|
|
rs369762002 CA332384235 |
18 | D>A | No |
ClinGen Ensembl |
|
|
CA10466644 rs770376710 |
18 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 18 | D>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs377498487 CA332384237 |
19 | S>N | No |
ClinGen ESP TOPMed |
|
|
CA413970430 rs1357700461 |
20 | P>A | No |
ClinGen gnomAD |
|
|
rs1302086878 CA413970439 |
21 | A>V | No |
ClinGen gnomAD |
|
|
rs769698929 CA10466647 CA332384240 |
22 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1328614742 CA413970474 |
26 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1375519933 CA413970480 |
27 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs749102763 CA10466649 |
31 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA413970543 COSM614146 rs761915511 |
36 | N>K | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs774461925 CA10466651 |
36 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10466653 COSM614145 rs540837899 |
37 | N>S | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs773693755 CA10466654 |
38 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA413970558 rs1238173623 |
39 | D>Y | No |
ClinGen gnomAD |
|
| TCGA novel | 40 | T>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs768861294 CA10466655 |
40 | T>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1327419078 CA413970576 |
42 | R>G | No |
ClinGen TOPMed |
|
|
rs1602292893 CA413970601 |
46 | L>V | No |
ClinGen Ensembl |
|
|
rs754143244 CA10466657 |
47 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1175028565 CA413970632 |
50 | K>R | No |
ClinGen gnomAD |
|
| TCGA novel | 51 | K>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA413970648 rs1378000023 |
52 | K>R | No |
ClinGen gnomAD |
|
| TCGA novel | 52 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA413970701 rs1470876065 |
60 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1433038545 CA413970709 |
61 | S>A | No |
ClinGen TOPMed |
|
|
CA10466660 rs763760276 |
62 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs375358921 CA10466661 |
63 | K>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs780908528 CA10466663 |
66 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs368249504 CA10466665 |
70 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA413970788 rs1289021144 |
72 | H>R | No |
ClinGen gnomAD |
|
|
rs909656285 CA332384261 |
84 | Q>E | No |
ClinGen TOPMed |
|
|
rs937492052 CA332384263 |
85 | M>I | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 88 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA413970901 rs1235544011 |
88 | E>G | No |
ClinGen TOPMed |
|
|
CA413970907 rs1481285880 |
89 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
rs142605569 CA10466672 |
91 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10466673 rs144880993 |
94 | L>V | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1466100920 CA413970942 |
94 | L>W | No |
ClinGen gnomAD |
|
|
CA10466674 rs766822144 |
95 | L>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs2496945 CA332384269 |
96 | Q>R | No |
ClinGen Ensembl |
|
|
rs1336452261 CA413971007 |
103 | N>S | No |
ClinGen TOPMed |
|
| TCGA novel | 107 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1312288646 CA413971036 |
108 | I>L | No |
ClinGen TOPMed |
|
|
CA413971054 rs1602292984 |
110 | P>L | No |
ClinGen Ensembl |
|
|
CA413971059 rs1253469101 |
111 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1347061100 CA413971082 |
114 | Q>R | No |
ClinGen gnomAD |
|
|
rs751212646 COSM614140 CA10466678 |
116 | R>H | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs756927449 CA10466679 |
118 | N>K | No |
ClinGen ExAC gnomAD |
|
|
COSM1469821 CA413971114 rs1341625302 |
119 | D>N | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
| TCGA novel | 120 | P>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA413971129 rs1216841783 |
121 | I>V | No |
ClinGen gnomAD |
|
|
CA413971139 rs1362307427 |
122 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1399935693 CA413971136 |
122 | I>V | No |
ClinGen TOPMed |
|
|
rs755711518 CA10466683 |
125 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs755711518 CA10466682 |
125 | K>T | No |
ClinGen ExAC gnomAD |
|
|
rs141862335 COSM3708748 CA10466684 |
126 | T>M | liver [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1470439903 CA413971174 |
128 | K>E | No |
ClinGen TOPMed |
|
|
CA413971181 rs1487450888 |
129 | D>N | No |
ClinGen gnomAD |
|
| TCGA novel | 130 | A>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1569308028 CA413971239 |
137 | S>N | No |
ClinGen Ensembl |
|
|
COSM1315720 CA10466687 rs747975762 |
139 | E>K | Variant assessed as Somatic; 0.0 impact. urinary_tract [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA413971258 rs1411613499 |
140 | A>P | No |
ClinGen gnomAD |
|
|
rs1422708039 CA413971262 |
140 | A>V | No |
ClinGen gnomAD |
|
|
rs1159265872 CA413971263 |
141 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
rs773755289 CA10466689 |
142 | V>A | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA413971279 rs1215398880 |
143 | P>L | No |
ClinGen TOPMed |
|
|
rs763411683 CA10466691 |
145 | K>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA413971298 TCGA novel rs1602293047 |
146 | S>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
NCI-TCGA ClinGen Ensembl |
|
CA10466694 rs770190881 |
149 | S>G | No |
ClinGen ExAC gnomAD |
|
|
COSM1558693 CA413971320 rs1379139812 |
149 | S>N | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA413971329 rs1225992105 |
150 | G>A | No |
ClinGen TOPMed |
|
| TCGA novel | 151 | P>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA413971355 rs1317283937 |
153 | N>S | No |
ClinGen gnomAD |
|
|
rs775650661 CA10466695 |
156 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767233454 CA10466697 |
159 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs763392229 CA10466696 |
159 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 166 | Q>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA413971466 rs1447352316 |
166 | Q>P | No |
ClinGen gnomAD |
|
|
CA413971477 rs1217742625 |
167 | M>I | No |
ClinGen gnomAD |
|
|
CA10466698 rs750133801 |
170 | E>A | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 172 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1472443593 CA413971523 |
172 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1313748735 CA413971543 |
175 | P>A | No |
ClinGen TOPMed |
|
|
CA10466699 rs367945077 |
175 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1319960098 CA413971570 COSM368256 |
178 | A>D | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed |
| TCGA novel | 178 | A>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs753807305 CA10466701 |
179 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA413971594 rs1380585236 |
181 | Q>E | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 183 | L>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs754800415 COSM1469824 CA10466702 |
185 | G>R | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1449417206 CA413971670 |
190 | K>R | No |
ClinGen gnomAD |
|
|
rs778919460 CA10466703 |
191 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA413971705 rs1185599182 |
195 | V>F | No |
ClinGen TOPMed |
|
|
VAR_017867 rs2290380 CA10466706 |
197 | V>I | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA413971725 rs1332583401 |
198 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA332384306 COSM107365 rs145641550 |
199 | S>F | skin [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA10466708 rs770986994 |
200 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781197938 CA10466709 |
200 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781197938 CA413971734 |
200 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA413971739 rs1204225534 |
201 | S>C | No |
ClinGen gnomAD |
|
|
CA332384310 rs1039543397 |
201 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1244960748 CA413971751 |
203 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 203 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10466710 rs745965870 |
205 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA10466711 rs770143319 |
206 | V>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs568525633 CA332384313 |
207 | S>T | No |
ClinGen Ensembl |
|
|
CA332384315 rs866571370 |
208 | P>S | No |
ClinGen Ensembl |
|
| TCGA novel | 209 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs776091366 CA10466712 |
211 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
CA332384317 rs34824030 |
211 | H>Y | No |
ClinGen Ensembl |
|
|
CA10466713 rs149582592 |
212 | A>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs149582592 CA413971802 |
212 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs772980516 CA413971847 |
218 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA10466716 rs767270814 |
219 | L>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA413971861 rs1179194729 |
221 | V>I | No |
ClinGen gnomAD |
|
|
rs765044031 CA10466720 |
222 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1337704759 CA413971874 |
223 | A>T | No |
ClinGen gnomAD |
|
|
rs1434595254 CA413971883 |
224 | A>V | No |
ClinGen gnomAD |
|
|
rs901052648 CA332384325 |
226 | Q>R | No |
ClinGen TOPMed |
|
|
CA332384327 rs34058937 |
227 | R>T | No |
ClinGen Ensembl |
|
|
CA413971924 rs1223948242 |
230 | E>D | No |
ClinGen gnomAD |
|
| TCGA novel | 230 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 232 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10466725 rs757405921 |
233 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10466726 rs781491795 |
235 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs756251143 CA10466728 |
238 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA10466729 rs756251143 |
238 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752282901 CA10466730 |
241 | P>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs369523286 CA10466731 |
242 | P>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
No associated diseases with Q8IUE1
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8MIB7 | TGIF2LX | Homeobox protein TGIF2LX | Pan troglodytes (Chimpanzee) | PR |
| P40424 | PBX1 | Pre-B-cell leukemia transcription factor 1 | Homo sapiens (Human) | PR |
| P40425 | PBX2 | Pre-B-cell leukemia transcription factor 2 | Homo sapiens (Human) | PR |
| O14770 | MEIS2 | Homeobox protein Meis2 | Homo sapiens (Human) | EV |
| Q99687 | MEIS3 | Homeobox protein Meis3 | Homo sapiens (Human) | SS |
| O00470 | MEIS1 | Homeobox protein Meis1 | Homo sapiens (Human) | SS |
| A8K0S8 | MEIS3P2 | Putative homeobox protein Meis3-like 2 | Homo sapiens (Human) | PR |
| A6NDR6 | MEIS3P1 | Putative homeobox protein Meis3-like 1 | Homo sapiens (Human) | PR |
| P97368 | Meis3 | Homeobox protein Meis3 | Mus musculus (Mouse) | PR |
| P97367 | Meis2 | Homeobox protein Meis2 | Mus musculus (Mouse) | SS |
| Q75LX7 | OSH10 | Homeobox protein knotted-1-like 4 | Oryza sativa subsp japonica (Rice) | PR |
| Q75LX9 | Os03g0673500 | Putative homeobox protein knotted-1-like 5 | Oryza sativa subsp japonica (Rice) | PR |
| Q1PFD1 | BLH11 | BEL1-like homeodomain protein 11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIW1 | BLH7 | BEL1-like homeodomain protein 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEAAADGPAE | TQSPVEKDSP | AKTQSPAQDT | SIMSRNNADT | GRVLALPEHK | KKRKGNLPAE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SVKILRDWMY | KHRFKAYPSE | EEKQMLSEKT | NLSLLQISNW | FINARRRILP | DMLQQRRNDP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IIGHKTGKDA | HATHLQSTEA | SVPAKSGPSG | PDNVQSLPLW | PLPKGQMSRE | KQPDPESAPS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QKLTGIAQPK | KKVKVSVTSP | SSPELVSPEE | HADFSSFLLL | VDAAVQRAAE | LELEKKQEPN |
| P |