Q8IU85
Gene name |
CAMK1D (CAMKID) |
Protein name |
Calcium/calmodulin-dependent protein kinase type 1D |
Names |
CaM kinase I delta, CaM kinase ID, CaM-KI delta, CaMKI delta, CaMKID, CaMKI-like protein kinase, CKLiK |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:57118 |
EC number |
2.7.11.17: Protein-serine/threonine kinases |
Protein Class |
|
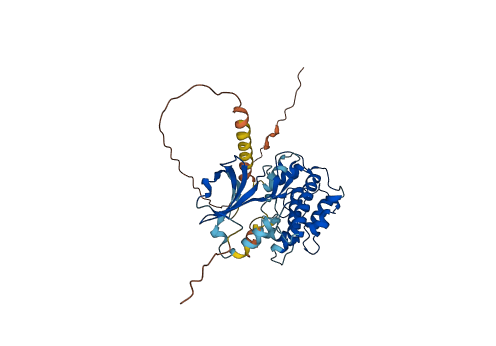
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
23-279 (Protein kinase domain) |
Relief mechanism |
Ligand binding |
Assay |
|
Accessory elements
164-186 (Activation loop from InterPro)
Target domain |
23-279 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yuan T et al. (2004) "Spectroscopic characterization of the calmodulin-binding and autoinhibitory domains of calcium/calmodulin-dependent protein kinase I", Archives of biochemistry and biophysics, 421, 192-206
- Goldberg J et al. (1996) "Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I", Cell, 84, 875-87
- Stull JT et al. (2011) "Myosin light chain kinase and the role of myosin light chain phosphorylation in skeletal muscle", Archives of biochemistry and biophysics, 510, 120-8
- Rellos P et al. (2010) "Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation", PLoS biology, 8, e1000426
- Akita T et al. (2018) "De novo variants in CAMK2A and CAMK2B cause neurodevelopmental disorders", Annals of clinical and translational neurology, 5, 280-296
- Yang E et al. (1999) "Structural examination of autoregulation of multifunctional calcium/calmodulin-dependent protein kinase II", The Journal of biological chemistry, 274, 26199-208
- Shohat G et al. (2002) "The DAP-kinase family of proteins: study of a novel group of calcium-regulated death-promoting kinases", Biochimica et biophysica acta, 1600, 45-50
- Cohen O et al. (1997) "DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity", The EMBO journal, 16, 998-1008
- Shohat G et al. (2001) "The pro-apoptotic function of death-associated protein kinase is controlled by a unique inhibitory autophosphorylation-based mechanism", The Journal of biological chemistry, 276, 47460-7
- Shani G et al. (2001) "Autophosphorylation restrains the apoptotic activity of DRP-1 kinase by controlling dimerization and calmodulin binding", The EMBO journal, 20, 1099-113
- Inbal B et al. (2000) "Death-associated protein kinase-related protein 1, a novel serine/threonine kinase involved in apoptosis", Molecular and cellular biology, 20, 1044-54
- Cheng L et al. (2022) "DCLK1 autoinhibition and activation in tumorigenesis", Innovation (Cambridge (Mass.)), 3, 100191
Autoinhibited structure

Activated structure

8 structures for Q8IU85
190 variants for Q8IU85
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA376217195 rs1343826343 |
5 | N>K | No |
ClinGen gnomAD |
|
|
CA5409759 rs772487155 |
8 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1281713706 CA376217219 |
9 | S>G | No |
ClinGen TOPMed |
|
|
rs1039929066 CA203120491 |
9 | S>I | No |
ClinGen TOPMed |
|
|
CA376217230 rs1212733154 |
10 | S>F | No |
ClinGen gnomAD |
|
|
CA376217270 rs1206974177 |
16 | A>T | No |
ClinGen gnomAD |
|
|
rs898711764 CA203120492 |
18 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs760852775 CA5409761 |
20 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs771123508 CA5409762 |
26 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs759616291 CA5409790 |
33 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA5409794 COSM915804 rs763942938 |
36 | E>K | large_intestine skin endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs370966780 CA5409796 |
41 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA376218450 rs1394762283 |
42 | E>A | No |
ClinGen TOPMed |
|
|
rs749966956 CA203146227 |
46 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs749966956 CA5409798 |
46 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA376218540 rs1254573817 |
49 | F>Y | No |
ClinGen gnomAD |
|
|
rs1174614968 CA376218561 |
51 | V>L | No |
ClinGen TOPMed |
|
|
CA5409801 rs753517881 |
56 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA203146228 rs980794056 |
57 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs758758416 CA5409802 |
58 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5409806 rs781753117 |
61 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA203146231 rs781753117 |
61 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA5409808 rs769900753 |
64 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs34194224 VAR_040599 RCV000972596 CA5409809 |
66 | I>M | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA376218775 rs1233023358 |
66 | I>T | No |
ClinGen TOPMed |
|
|
CA203146232 rs972745162 |
67 | E>G | No |
ClinGen Ensembl |
|
|
CA376218838 rs1227393204 |
71 | A>T | No |
ClinGen gnomAD |
|
|
COSM181067 CA5409811 rs768486499 |
72 | V>I | large_intestine endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA5409813 rs761555171 |
75 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767447307 CA5409814 |
75 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1163658647 CA376217450 |
82 | V>I | No |
ClinGen gnomAD |
|
|
CA5409858 rs771919287 |
83 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA376217484 rs1317883329 |
87 | I>V | No |
ClinGen gnomAD |
|
|
rs1435005582 CA376217519 |
92 | N>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1364493322 CA376217555 |
97 | V>I | No |
ClinGen gnomAD |
|
|
rs267602410 CA202596909 |
101 | V>M | No |
ClinGen Ensembl |
|
|
CA376011341 rs1588895401 COSM3396980 |
103 | G>S | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs774092855 CA5409903 |
108 | D>A | No |
ClinGen ExAC gnomAD |
|
|
CA376011826 rs1331020416 |
109 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs776992169 CA5409906 |
110 | I>M | No |
ClinGen ExAC gnomAD |
|
|
CA376011880 rs1435788333 |
112 | E>K | No |
ClinGen gnomAD |
|
|
rs759827266 CA5409907 |
113 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs765761653 CA5409908 |
113 | K>R | No |
ClinGen ExAC TOPMed |
|
|
CA376011957 rs1588895447 |
115 | F>V | No |
ClinGen Ensembl |
|
|
CA202596948 rs867043228 |
121 | A>G | No |
ClinGen Ensembl |
|
|
COSM1247274 rs143417427 CA5409911 |
126 | R>C | oesophagus [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs1322015233 CA376012345 |
126 | R>H | No |
ClinGen gnomAD |
|
|
rs1287192477 CA376012619 |
132 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
rs750276790 CA5409915 |
133 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs750276790 CA376012656 |
133 | Y>F | No |
ClinGen ExAC gnomAD |
|
|
rs767783414 CA5409914 |
133 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
CA376012774 rs1455952139 |
138 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
CA5409917 rs780191189 |
140 | I>V | No |
ClinGen ExAC gnomAD |
|
|
COSM1297054 CA5409942 rs752176853 |
148 | E>K | urinary_tract [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA376015371 rs1442102703 |
149 | N>D | No |
ClinGen gnomAD |
|
|
CA202602939 rs267602411 |
151 | L>S | No |
ClinGen Ensembl |
|
|
CA5409947 rs780300077 |
153 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs1300903656 CA376015478 |
155 | Q>K | No |
ClinGen TOPMed gnomAD |
|
|
rs769078416 CA5409949 |
156 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA376015494 rs1401203311 |
156 | D>N | No |
ClinGen gnomAD |
|
|
CA376015609 rs1314361890 |
163 | I>N | No |
ClinGen gnomAD |
|
|
CA5409950 rs774874665 |
164 | S>I | No |
ClinGen ExAC gnomAD |
|
|
rs1184966710 CA376015692 |
167 | G>* | No |
ClinGen TOPMed |
|
|
CA376015773 rs1318121764 |
171 | M>T | No |
ClinGen gnomAD |
|
|
rs761850306 CA5409951 |
172 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA202603007 rs1001126310 |
173 | G>D | No |
ClinGen gnomAD |
|
|
CA5409952 rs772247725 |
173 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1001126310 CA376015804 |
173 | G>V | No |
ClinGen gnomAD |
|
|
rs1242186155 CA376015828 |
175 | G>E | No |
ClinGen TOPMed |
|
|
CA202603010 rs368615989 |
176 | D>V | No |
ClinGen ESP |
|
|
rs773637002 CA5409953 |
181 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA376016060 rs1286796548 |
187 | Y>C | No |
ClinGen TOPMed |
|
|
rs1350669979 CA376016088 |
189 | A>T | No |
ClinGen TOPMed |
|
|
CA5409977 rs759296135 |
190 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs762894154 CA5409980 |
194 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5409981 rs762894154 |
194 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA376020752 rs1296563583 |
194 | A>V | No |
ClinGen gnomAD |
|
|
rs560397402 CA202613625 |
199 | S>G | No |
ClinGen Ensembl |
|
|
rs1427849008 CA376020850 |
201 | A>V | No |
ClinGen Ensembl |
|
|
CA376020860 rs1344585922 |
202 | V>I | No |
ClinGen TOPMed |
|
|
CA376021151 rs1236594782 |
212 | Y>C | No |
ClinGen gnomAD |
|
|
CA376025226 rs1410471308 |
215 | L>R | No |
ClinGen gnomAD |
|
|
rs1160339552 CA376025236 |
217 | G>S | No |
ClinGen gnomAD |
|
|
rs547155036 CA5410007 |
218 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
CA5410009 rs758785829 |
219 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1588959590 CA376025272 |
222 | Y>F | No |
ClinGen Ensembl |
|
|
rs191753449 CA202630942 |
223 | D>Y | No |
ClinGen Ensembl |
|
|
rs181884337 CA202630952 |
224 | E>* | No |
ClinGen Ensembl |
|
|
CA376025297 rs1185940422 |
225 | N>K | No |
ClinGen TOPMed |
|
|
rs1416718650 CA376025313 |
228 | K>E | No |
ClinGen gnomAD |
|
|
rs778470103 CA5410010 |
228 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757343559 CA5410012 |
229 | L>I | No |
ClinGen ExAC |
|
|
COSM1346711 CA376025471 rs1257797457 |
236 | A>V | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA376025479 rs1200525794 |
237 | E>K | No |
ClinGen TOPMed |
|
|
rs770090758 CA5410015 |
239 | E>V | No |
ClinGen ExAC gnomAD |
|
|
CA376025537 rs1307994315 |
241 | D>N | No |
ClinGen TOPMed |
|
|
rs768410577 CA5410018 |
242 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs780203500 CA5410016 |
242 | S>P | No |
ClinGen ExAC gnomAD |
|
|
rs780203500 CA5410017 |
242 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs761796913 CA5410020 |
250 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762036652 CA5410048 |
254 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA5410049 rs201325490 |
255 | F>L | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs202177332 CA5410050 |
257 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs200489408 CA5410052 |
257 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA5410051 COSM3414811 rs202177332 |
257 | R>W | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs753859699 CA376026141 |
258 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5410054 rs755199191 |
260 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA5410055 rs779065638 |
262 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs747995993 CA5410056 |
262 | K>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs973947060 CA202633259 |
264 | P>L | No |
ClinGen TOPMed |
|
|
rs777804626 CA5410058 |
265 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs746912134 CA5410059 |
266 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1317342449 CA592069652 |
268 | Y>* | No |
ClinGen gnomAD |
|
|
COSM73997 rs745306887 CA5410062 |
269 | T>M | ovary [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1367464554 CA376026226 |
271 | E>K | No |
ClinGen TOPMed |
|
|
rs1273177100 CA376026248 |
274 | A>T | No |
ClinGen TOPMed |
|
|
CA5410065 rs185727180 |
275 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs779096797 CA5410064 |
275 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA376026718 rs912179319 |
280 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA202640138 rs912179319 |
280 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1324031455 CA376026726 |
281 | G>A | No |
ClinGen gnomAD |
|
|
CA202640139 rs142495899 |
282 | D>E | No |
ClinGen ESP |
|
|
CA202640140 rs991717263 |
286 | N>S | No |
ClinGen Ensembl |
|
|
CA5410095 rs138858875 |
290 | H>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1390244057 CA376026790 |
291 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
CA5410098 rs757099962 |
293 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA5410100 COSM98870 rs750385169 |
295 | A>T | stomach [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA5410101 COSM181128 rs201893185 |
298 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA376026903 rs1282662078 |
306 | W>* | No |
ClinGen TOPMed gnomAD |
|
|
rs1282662078 CA376026904 |
306 | W>C | No |
ClinGen TOPMed gnomAD |
|
|
rs748766971 CA5410103 |
307 | R>K | No |
ClinGen ExAC gnomAD |
|
|
CA376010172 rs186526367 |
308 | Q>E | No |
ClinGen TOPMed |
|
|
CA202614243 rs186526367 |
308 | Q>K | No |
ClinGen TOPMed |
|
|
rs868730254 CA202614264 |
312 | A>V | No |
ClinGen Ensembl |
|
|
rs1381853441 CA376010238 |
313 | T>A | No |
ClinGen gnomAD |
|
|
CA5410121 COSM1346717 rs754487170 |
313 | T>M | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA376010254 rs1231487507 |
314 | A>D | No |
ClinGen gnomAD |
|
|
CA5410125 rs139838737 |
315 | V>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs770233179 CA5410127 |
316 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1186753386 CA376010286 |
317 | R>I | No |
ClinGen gnomAD |
|
|
rs1588973087 CA376010347 |
323 | H>P | No |
ClinGen Ensembl |
|
|
rs200236740 CA5410129 |
323 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5410131 rs774775892 |
325 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA376010371 rs1163587540 |
327 | S>G | No |
ClinGen gnomAD |
|
|
rs1232161533 CA376010440 |
332 | N>S | No |
ClinGen TOPMed |
|
|
CA376010464 rs1462424421 |
334 | S>G | No |
ClinGen gnomAD |
|
|
CA5410133 rs767783056 |
336 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA5410135 rs760472229 |
337 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs565781008 CA5410136 |
337 | S>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA5410137 rs753775858 |
338 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754511728 CA5410138 |
338 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs753775858 CA202614418 |
338 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA376010545 rs1292715686 |
340 | S>G | No |
ClinGen gnomAD |
|
|
rs764713214 CA5410139 |
340 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs868098610 CA202614453 |
342 | A>V | No |
ClinGen Ensembl |
|
|
rs1281990165 CA376010581 |
344 | Q>R | No |
ClinGen gnomAD |
|
|
rs777362227 CA5410142 |
346 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs188569446 CA5410176 |
348 | L>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775381754 CA5410177 |
349 | A>E | No |
ClinGen ExAC gnomAD |
|
|
CA5410178 rs762520208 |
350 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs867488049 CA202618326 |
351 | S>P | No |
ClinGen Ensembl |
|
|
CA5410179 rs763584729 |
352 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs867692056 CA376012435 |
356 | F>L | No |
ClinGen gnomAD |
|
|
rs1003034685 CA202618349 |
357 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
rs1166288705 CA376012505 |
358 | S>Y | No |
ClinGen gnomAD |
|
|
CA5410183 rs148446769 |
360 | S>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5410186 rs779605714 |
361 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA202618430 rs896113789 |
362 | G>R | No |
ClinGen gnomAD |
|
|
CA5410190 rs777921875 |
363 | V>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5410189 rs777921875 |
363 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771212787 CA5410191 |
366 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA202618446 rs886676558 |
367 | G>E | No |
ClinGen TOPMed gnomAD |
|
|
CA376012793 rs1219887504 |
368 | A>T | No |
ClinGen gnomAD |
|
|
CA5410195 rs775467364 |
369 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5410194 rs200052874 |
369 | E>K | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA376012809 rs200052874 |
369 | E>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs768414434 CA5410197 |
370 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5410196 rs762894114 |
370 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA376012872 rs1160301011 |
372 | P>L | No |
ClinGen gnomAD |
|
|
rs773947005 CA5410198 |
372 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs371362652 CA5410201 |
375 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5410202 rs760043332 |
376 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA5410203 rs765682396 |
378 | T>M | No |
ClinGen ExAC gnomAD |
|
|
rs753052899 CA5410204 |
380 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA5410206 rs374664435 |
381 | H>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA5410205 rs145491457 |
381 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1290867546 CA376013050 |
384 | S>R | No |
ClinGen gnomAD |
|
|
CA5410207 rs751678897 |
384 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA202618593 rs966639211 |
385 | K>N | No |
ClinGen Ensembl |
|
|
rs933186919 CA202618596 |
386 | K>C | No |
ClinGen TOPMed |
No associated diseases with Q8IU85
No regional properties for Q8IU85
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q8IU85 | |||
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.17 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein kinase activity | Calmodulin-dependent catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; and ATP + a protein threonine = ADP + protein threonine phosphate. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| nervous system development | The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of CREB transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of the transcription factor CREB. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| positive regulation of neutrophil chemotaxis | Any process that increases the frequency, rate, or extent of neutrophil chemotaxis. Neutrophil chemotaxis is the directed movement of a neutrophil cell, the most numerous polymorphonuclear leukocyte found in the blood, in response to an external stimulus, usually an infection or wounding. |
| positive regulation of phagocytosis | Any process that activates or increases the frequency, rate or extent of phagocytosis. |
| positive regulation of respiratory burst | Any process that increases the rate frequency or extent of a phase of elevated metabolic activity, during which oxygen consumption increases; this leads to the production, by an NADH dependent system, of hydrogen peroxide (H2O2), superoxide anions and hydroxyl radicals. |
| regulation of dendrite development | Any process that modulates the frequency, rate or extent of dendrite development. |
| regulation of granulocyte chemotaxis | Any process that modulates the rate, frequency or extent of granulocyte chemotaxis. Granulocyte chemotaxis is the movement of a granulocyte in response to an external stimulus. |
106 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A4IFM7 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Bos taurus (Bovine) | SS |
| Q0V7M1 | PSKH1 | Serine/threonine-protein kinase H1 | Bos taurus (Bovine) | SS |
| Q0VD22 | STK33 | Serine/threonine-protein kinase 33 | Bos taurus (Bovine) | PR |
| O15075 | DCLK1 | Serine/threonine-protein kinase DCLK1 | Homo sapiens (Human) | EV |
| Q9UIK4 | DAPK2 | Death-associated protein kinase 2 | Homo sapiens (Human) | EV |
| O94768 | STK17B | Serine/threonine-protein kinase 17B | Homo sapiens (Human) | PR |
| Q16566 | CAMK4 | Calcium/calmodulin-dependent protein kinase type IV | Homo sapiens (Human) | SS |
| Q8N568 | DCLK2 | Serine/threonine-protein kinase DCLK2 | Homo sapiens (Human) | PR |
| Q9UEE5 | STK17A | Serine/threonine-protein kinase 17A | Homo sapiens (Human) | SS |
| O75962 | TRIO | Triple functional domain protein | Homo sapiens (Human) | EV |
| Q8WWF8 | CAPSL | Calcyphosin-like protein | Homo sapiens (Human) | PR |
| P53355 | DAPK1 | Death-associated protein kinase 1 | Homo sapiens (Human) | EV |
| Q9BYT3 | STK33 | Serine/threonine-protein kinase 33 | Homo sapiens (Human) | PR |
| O43293 | DAPK3 | Death-associated protein kinase 3 | Homo sapiens (Human) | PR |
| Q16816 | PHKG1 | Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform | Homo sapiens (Human) | PR |
| P15735 | PHKG2 | Phosphorylase b kinase gamma catalytic chain, liver/testis isoform | Homo sapiens (Human) | PR |
| Q13554 | CAMK2B | Calcium/calmodulin-dependent protein kinase type II subunit beta | Homo sapiens (Human) | EV |
| Q9UQM7 | CAMK2A | Calcium/calmodulin-dependent protein kinase type II subunit alpha | Homo sapiens (Human) | EV |
| Q13555 | CAMK2G | Calcium/calmodulin-dependent protein kinase type II subunit gamma | Homo sapiens (Human) | EV |
| Q13557 | CAMK2D | Calcium/calmodulin-dependent protein kinase type II subunit delta | Homo sapiens (Human) | EV |
| Q9H1R3 | MYLK2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Homo sapiens (Human) | EV |
| Q32MK0 | MYLK3 | Myosin light chain kinase 3 | Homo sapiens (Human) | SS |
| Q86YV6 | MYLK4 | Myosin light chain kinase family member 4 | Homo sapiens (Human) | SS |
| P11801 | PSKH1 | Serine/threonine-protein kinase H1 | Homo sapiens (Human) | SS |
| Q6P2M8 | PNCK | Calcium/calmodulin-dependent protein kinase type 1B | Homo sapiens (Human) | SS |
| Q96NX5 | CAMK1G | Calcium/calmodulin-dependent protein kinase type 1G | Homo sapiens (Human) | SS |
| Q8NCB2 | CAMKV | CaM kinase-like vesicle-associated protein | Homo sapiens (Human) | SS |
| Q14012 | CAMK1 | Calcium/calmodulin-dependent protein kinase type 1 | Homo sapiens (Human) | EV |
| Q9QYK9 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Mus musculus (Mouse) | PR |
| Q91VB2 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Mus musculus (Mouse) | SS |
| Q8BG48 | Stk17b | Serine/threonine-protein kinase 17B | Mus musculus (Mouse) | PR |
| Q8VCR8 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Mus musculus (Mouse) | SS |
| Q6PGN3 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Mus musculus (Mouse) | PR |
| Q91YS8 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Mus musculus (Mouse) | SS |
| Q3UIZ8 | Mylk3 | Myosin light chain kinase 3 | Mus musculus (Mouse) | SS |
| Q8VDF3 | Dapk2 | Death-associated protein kinase 2 | Mus musculus (Mouse) | EV |
| Q6P8Y1 | Capsl | Calcyphosin-like protein | Mus musculus (Mouse) | PR |
| Q3UHL1 | Camkv | CaM kinase-like vesicle-associated protein | Mus musculus (Mouse) | SS |
| P08414 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Mus musculus (Mouse) | SS |
| Q9JLM8 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Mus musculus (Mouse) | SS |
| Q91YA2 | Pskh1 | Serine/threonine-protein kinase H1 | Mus musculus (Mouse) | SS |
| Q80YE7 | Dapk1 | Death-associated protein kinase 1 | Mus musculus (Mouse) | SS |
| Q62407 | Speg | Striated muscle-specific serine/threonine-protein kinase | Mus musculus (Mouse) | SS |
| Q0KL02 | Trio | Triple functional domain protein | Mus musculus (Mouse) | SS |
| Q924X7 | Stk33 | Serine/threonine-protein kinase 33 | Mus musculus (Mouse) | PR |
| O54784 | Dapk3 | Death-associated protein kinase 3 | Mus musculus (Mouse) | PR |
| Q8BW96 | Camk1d | Calcium/calmodulin-dependent protein kinase type 1D | Mus musculus (Mouse) | SS |
| Q7TNJ7 | Camk1g | Calcium/calmodulin-dependent protein kinase type 1G | Rattus norvegicus (Rat) | SS |
| P20689 | Mylk2 | Myosin light chain kinase 2, skeletal/cardiac muscle | Rattus norvegicus (Rat) | SS |
| F1M0Z1 | Trio | Triple functional domain protein | Rattus norvegicus (Rat) | SS |
| Q91XS8 | Stk17b | Serine/threonine-protein kinase 17B | Rattus norvegicus (Rat) | PR |
| O08875 | Dclk1 | Serine/threonine-protein kinase DCLK1 | Rattus norvegicus (Rat) | PR |
| E9PT87 | Mylk3 | Myosin light chain kinase 3 | Rattus norvegicus (Rat) | SS |
| O70150 | Pnck | Calcium/calmodulin-dependent protein kinase type 1B | Rattus norvegicus (Rat) | PR |
| Q63092 | Camkv | CaM kinase-like vesicle-associated protein | Rattus norvegicus (Rat) | SS |
| Q63638 | Speg | Striated muscle-specific serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| O88764 | Dapk3 | Death-associated protein kinase 3 | Rattus norvegicus (Rat) | PR |
| P13234 | Camk4 | Calcium/calmodulin-dependent protein kinase type IV | Rattus norvegicus (Rat) | SS |
| Q63450 | Camk1 | Calcium/calmodulin-dependent protein kinase type 1 | Rattus norvegicus (Rat) | EV |
| P97924 | Kalrn | Kalirin | Rattus norvegicus (Rat) | SS |
| Q5MPA9 | Dclk2 | Serine/threonine-protein kinase DCLK2 | Rattus norvegicus (Rat) | PR |
| Q6AVM3 | CCAMK | Calcium and calcium/calmodulin-dependent serine/threonine-protein kinase | Oryza sativa subsp japonica (Rice) | PR |
| P53682 | CPK23 | Calcium-dependent protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| P53684 | CPK7 | Calcium-dependent protein kinase 7 | Oryza sativa subsp japonica (Rice) | PR |
| Q5VQQ5 | CPK2 | Calcium-dependent protein kinase 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6I5I8 | CPK16 | Calcium-dependent protein kinase 16 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6K968 | CPK6 | Calcium-dependent protein kinase 6 | Oryza sativa subsp. japonica (Rice) | SS |
| Q75GE8 | CPK8 | Calcium-dependent protein kinase 8 | Oryza sativa subsp. japonica (Rice) | SS |
| Q84SL0 | CPK20 | Calcium-dependent protein kinase 20 | Oryza sativa subsp. japonica (Rice) | SS |
| Q8LPZ7 | CPK3 | Calcium-dependent protein kinase 3 | Oryza sativa subsp. japonica (Rice) | SS |
| O44997 | dapk-1 | Death-associated protein kinase dapk-1 | Caenorhabditis elegans | SS |
| Q95QC4 | zyg-8 | Serine/threonine-protein kinase zyg-8 | Caenorhabditis elegans | PR |
| Q9TXJ0 | cmk-1 | Calcium/calmodulin-dependent protein kinase type 1 | Caenorhabditis elegans | PR |
| P28583 | Calcium-dependent protein kinase SK5 | Glycine max (Soybean) (Glycine hispida) | PR | |
| Q9M9V8 | CPK10 | Calcium-dependent protein kinase 10 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FX86 | CRK8 | CDPK-related kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SSF8 | CPK30 | Calcium-dependent protein kinase 30 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIQ7 | CPK24 | Calcium-dependent protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93759 | GK-1 | Calcium-dependent protein kinase 14 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8S8S2 | LPEAT2 | Lysophospholipid acyltransferase LPEAT2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LJL9 | CRK2 | CDPK-related kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SG12 | CRK6 | CDPK-related kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SCS2 | CRK5 | CDPK-related kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8W4I7 | CPK13 | Calcium-dependent protein kinase 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NLQ6 | CPK32 | Calcium-dependent protein kinase 32 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38873 | CPK7 | Calcium-dependent protein kinase 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42438 | CPK8 | Calcium-dependent protein kinase 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q06850 | CPK1 | Calcium-dependent protein kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38869 | CPK4 | Calcium-dependent protein kinase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38870 | CPK2 | Calcium-dependent protein kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38872 | CPK6 | Calcium-dependent protein kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39016 | CPK11 | Calcium-dependent protein kinase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E9C0 | CPK34 | Calcium-dependent protein kinase 34 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q42396 | CPK12 | Calcium-dependent protein kinase 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42479 | CPK3 | Calcium-dependent protein kinase 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q7XJR9 | CPK16 | Calcium-dependent protein kinase 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FMP5 | CPK17 | Calcium-dependent protein kinase 17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJ61 | CPK25 | Calcium-dependent protein kinase 25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZSA4 | CPK27 | Calcium-dependent protein kinase 27 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZV15 | CPK20 | Calcium-dependent protein kinase 20 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q1PE17 | CPK18 | Calcium-dependent protein kinase 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FKW4 | CPK28 | Calcium-dependent protein kinase 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1LUA6 | trio | Triple functional domain protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A8C984 | mylk3 | Myosin light chain kinase 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q7SY49 | camkv | CaM kinase-like vesicle-associated protein | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MARENGESSS | SWKKQAEDIK | KIFEFKETLG | TGAFSEVVLA | EEKATGKLFA | VKCIPKKALK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GKESSIENEI | AVLRKIKHEN | IVALEDIYES | PNHLYLVMQL | VSGGELFDRI | VEKGFYTEKD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ASTLIRQVLD | AVYYLHRMGI | VHRDLKPENL | LYYSQDEESK | IMISDFGLSK | MEGKGDVMST |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ACGTPGYVAP | EVLAQKPYSK | AVDCWSIGVI | AYILLCGYPP | FYDENDSKLF | EQILKAEYEF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| DSPYWDDISD | SAKDFIRNLM | EKDPNKRYTC | EQAARHPWIA | GDTALNKNIH | ESVSAQIRKN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| FAKSKWRQAF | NATAVVRHMR | KLHLGSSLDS | SNASVSSSLS | LASQKDCLAP | STLCSFISSS |
| 370 | 380 | ||||
| SGVSGVGAER | RPRPTTVTAV | HSGSK |