Q8H1F2
Gene name |
RGS1 (At3g26090, MPE11.27, MPE11.28) |
Protein name |
Regulator of G-protein signaling 1 |
Names |
AtRGS1 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT3G26090 |
EC number |
|
Protein Class |
|
Descriptions
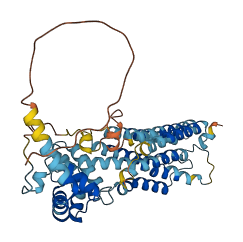
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q8H1F2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8H1F2-F1 | Predicted | AlphaFoldDB |
42 variants for Q8H1F2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH05919907 | 6 | A>S | No | 1000Genomes | |
| tmp_3_9532928_G_T | 16 | V>F | No | 1000Genomes | |
| ENSVATH05919908 | 27 | V>A | No | 1000Genomes | |
| ENSVATH02206875 | 44 | T>I | No | 1000Genomes | |
| ENSVATH05919915 | 48 | S>N | No | 1000Genomes | |
| tmp_3_9533643_C_T | 77 | T>M | No | 1000Genomes | |
| ENSVATH10999047 | 125 | S>L | No | 1000Genomes | |
| ENSVATH00351304 | 151 | D>Y | No | 1000Genomes | |
| tmp_3_9534155_A_C | 155 | M>L | No | 1000Genomes | |
| tmp_3_9534210_C_G | 173 | A>G | No | 1000Genomes | |
| tmp_3_9534218_G_T | 176 | A>S | No | 1000Genomes | |
| tmp_3_9534236_C_T | 182 | R>* | No | 1000Genomes | |
| tmp_3_9534308_G_A,C | 206 | V>I | No | 1000Genomes | |
| tmp_3_9534308_G_A,C | 206 | V>L | No | 1000Genomes | |
| ENSVATH00351305 | 210 | T>A | No | 1000Genomes | |
| ENSVATH02206879 | 211 | A>V | No | 1000Genomes | |
| ENSVATH05919926 | 221 | I>N | No | 1000Genomes | |
| ENSVATH05919928 | 242 | V>L | No | 1000Genomes | |
| ENSVATH10999054 | 269 | M>I | No | 1000Genomes | |
| ENSVATH00351307 | 298 | K>M | No | 1000Genomes | |
| ENSVATH10999085 | 306 | R>C | No | 1000Genomes | |
| tmp_3_9534697_A_T | 307 | H>L | No | 1000Genomes | |
| ENSVATH05919929 | 309 | F>L | No | 1000Genomes | |
| ENSVATH10999086 | 316 | C>S | No | 1000Genomes | |
| tmp_3_9534867_A_T | 338 | D>V | No | 1000Genomes | |
| tmp_3_9534872_A_G | 340 | I>V | No | 1000Genomes | |
| ENSVATH05919933 | 341 | R>K | No | 1000Genomes | |
| ENSVATH14014247 | 342 | R>T | No | 1000Genomes | |
| ENSVATH00351309 | 362 | L>V | No | 1000Genomes | |
| tmp_3_9535054_G_A | 371 | E>K | No | 1000Genomes | |
| ENSVATH05919938 | 391 | V>A | No | 1000Genomes | |
| ENSVATH14014249 | 409 | F>S | No | 1000Genomes | |
| ENSVATH05919940 | 415 | E>K | No | 1000Genomes | |
| ENSVATH00351311 | 419 | H>D | No | 1000Genomes | |
| ENSVATH05919941 | 419 | H>Q | No | 1000Genomes | |
| ENSVATH05919942 | 423 | H>Q | No | 1000Genomes | |
| ENSVATH14014250 | 424 | K>T | No | 1000Genomes | |
| ENSVATH10999090 | 446 | Q>L | No | 1000Genomes | |
| ENSVATH05919943 | 447 | E>D | No | 1000Genomes | |
| ENSVATH05919944 | 448 | H>R | No | 1000Genomes | |
| tmp_3_9535384_A_T | 452 | S>C | No | 1000Genomes | |
| ENSVATH10999091 | 454 | R>G | No | 1000Genomes |
No associated diseases with Q8H1F2
1 regional properties for Q8H1F2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | RGS domain | 295 - 413 | IPR016137 |
Functions
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| endosome membrane | The lipid bilayer surrounding an endosome. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein-coupled receptor activity | Combining with an extracellular signal and transmitting the signal across the membrane by activating an associated G-protein; promotes the exchange of GDP for GTP on the alpha subunit of a heterotrimeric G-protein complex. |
| GTPase activator activity | Binds to and increases the activity of a GTPase, an enzyme that catalyzes the hydrolysis of GTP. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| jasmonic acid mediated signaling pathway | The series of molecular signals mediated by jasmonic acid. |
| negative regulation of signal transduction | Any process that stops, prevents, or reduces the frequency, rate or extent of signal transduction. |
| positive regulation of GTPase activity | Any process that activates or increases the activity of a GTPase. |
| regulation of cell population proliferation | Any process that modulates the frequency, rate or extent of cell proliferation. |
| response to abscisic acid | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an abscisic acid stimulus. |
| response to glucose | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a glucose stimulus. |
| response to water deprivation | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a water deprivation stimulus, prolonged deprivation of water. |
| sugar mediated signaling pathway | The process in which a change in the level of a mono- or disaccharide such as glucose, fructose or sucrose triggers the expression of genes controlling metabolic and developmental processes. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3T0T8 | RGS5 | Regulator of G-protein signaling 5 | Bos taurus (Bovine) | PR |
| O46471 | RGS16 | Regulator of G-protein signaling 16 | Bos taurus (Bovine) | PR |
| O15539 | RGS5 | Regulator of G-protein signaling 5 | Homo sapiens (Human) | PR |
| Q18312 | rgs-3 | Regulator of G-protein signaling rgs-3 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASGCALHGG | CPSDYVAVAI | SVICFFVLLS | RSVLPCLIHK | APRTNSSSFW | IPVIQVISSF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NLLFSIMMSV | NLLRFRTKHW | WRYCYLWAVW | IEGPLGFGLL | MSCRITQAFQ | LYFIFVKKRL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PPVKSYIFLP | LVLLPWIFGA | AIIHATKPLN | DKCHMGLQWT | FPVAGLHALY | VLALIAFTRA |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VRHVEFRFDE | LRDLWKGILV | SATSIVIWVT | AFVLNEIHEE | ISWLQVASRF | VLLVTGGILV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VVFFSISSNQ | PLLSQISLKK | RQNFEFQRMG | QALGIPDSGL | LFRKEEFRPV | DPNEPLDKLL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LNKRFRHSFM | EFADSCYAGE | TLHFFEEVYE | HGKIPEDDSI | RRIYMARHIM | EKFIVAGAEM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ELNLSHKTRQ | EILTTQDLTH | TDLFKNALNE | VMQLIKMNLV | RDYWSSIYFI | KFKEEESCHE |
| 430 | 440 | 450 | |||
| AMHKEGYSFS | SPRLSSVQGS | DDPFYQEHMS | KSSRCSSPG |