Q8CCN1
Gene name |
Nlrp10 (Nalp10, Pynod) |
Protein name |
NACHT, LRR and PYD domains-containing protein 10 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:244202 |
EC number |
|
Protein Class |
|
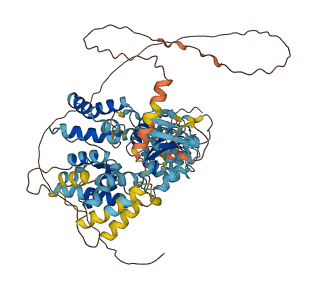
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q8CCN1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2DO9 | NMR | - | A | 1-102 | PDB |
| AF-Q8CCN1-F1 | Predicted | AlphaFoldDB |
37 variants for Q8CCN1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388940269 | 25 | F>S | No | EVA | |
| rs3388935161 | 37 | Q>R | No | EVA | |
| rs3388935195 | 55 | S>T | No | EVA | |
| rs3388943812 | 72 | R>T | No | EVA | |
| rs3388935353 | 99 | R>W | No | EVA | |
| rs215041372 | 103 | R>G | No | EVA | |
| rs3388935416 | 136 | C>* | No | EVA | |
| rs253128179 | 140 | E>Q | No | EVA | |
| rs32090088 | 144 | D>E | No | EVA | |
| rs3398370066 | 165 | V>T | No | EVA | |
| rs3388935193 | 172 | G>V | No | EVA | |
| rs3388943826 | 211 | P>A | No | EVA | |
| rs3388932586 | 232 | E>D | No | EVA | |
| rs258196731 | 258 | C>R | No | EVA | |
| rs3388921099 | 288 | M>I | No | EVA | |
| rs3398240721 | 293 | R>L | No | EVA | |
| rs3397981355 | 293 | R>Q | No | EVA | |
| rs225240175 | 394 | V>I | No | EVA | |
| rs243841194 | 439 | A>E | No | EVA | |
| rs243841194 | 439 | A>V | No | EVA | |
| rs242565842 | 507 | G>S | No | EVA | |
| rs3388943808 | 516 | C>* | No | EVA | |
| rs3388940226 | 517 | F>I | No | EVA | |
| rs32088966 | 547 | F>S | No | EVA | |
| rs3388940195 | 557 | L>H | No | EVA | |
| rs253330544 | 562 | Q>K | No | EVA | |
| rs231308858 | 585 | S>L | No | EVA | |
| rs3388935156 | 590 | F>V | No | EVA | |
| rs3388938549 | 596 | Q>L | No | EVA | |
| rs223593844 | 603 | D>G | No | EVA | |
| rs864270359 | 613 | S>T | No | EVA | |
| rs255899002 | 643 | N>D | No | EVA | |
| rs237441044 | 643 | N>S | No | EVA | |
| rs3388921116 | 657 | E>D | No | EVA | |
| rs3388935419 | 661 | L>F | No | EVA | |
| rs247583854 | 665 | G>E | No | EVA | |
| rs218440427 | 665 | G>R | No | EVA |
No associated diseases with Q8CCN1
Functions
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| extrinsic component of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to one of its surfaces, but not integrated into the hydrophobic region. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| adaptive immune response | An immune response mediated by cells expressing specific receptors for antigen produced through a somatic diversification process, and allowing for an enhanced secondary response to subsequent exposures to the same antigen (immunological memory). |
| defense response to fungus | Reactions triggered in response to the presence of a fungus that act to protect the cell or organism. |
| defense response to Gram-negative bacterium | Reactions triggered in response to the presence of a Gram-negative bacterium that act to protect the cell or organism. |
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| positive regulation of defense response to bacterium | Any process that activates or increases the frequency, rate or extent of defense response to bacterium. |
| positive regulation of inflammatory response | Any process that activates or increases the frequency, rate or extent of the inflammatory response. |
| positive regulation of interleukin-1 alpha production | Any process that activates or increases the frequency, rate, or extent of interleukin-1 alpha production. |
| positive regulation of interleukin-6 production | Any process that activates or increases the frequency, rate, or extent of interleukin-6 production. |
| positive regulation of interleukin-8 production | Any process that activates or increases the frequency, rate, or extent of interleukin-8 production. |
| positive regulation of T-helper 1 type immune response | Any process that activates or increases the frequency, rate, or extent of a T-helper 1 type immune response. |
| positive regulation of T-helper 17 type immune response | Any process that activates or increases the frequency, rate or extent of T-helper 17 type immune response. |
| regulation of inflammatory response | Any process that modulates the frequency, rate or extent of the inflammatory response, the immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q86W26 | NLRP10 | NACHT, LRR and PYD domains-containing protein 10 | Homo sapiens (Human) | PR |
| Q66X01 | Nlrp9c | NACHT, LRR and PYD domains-containing protein 9C | Mus musculus (Mouse) | PR |
| Q8R4B8 | Nlrp3 | NACHT, LRR and PYD domains-containing protein 3 | Mus musculus (Mouse) | SS |
| E9Q5R7 | Nlrp12 | NACHT, LRR and PYD domains-containing protein 12 | Mus musculus (Mouse) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MALARANSPQ | EALLWALNDL | EENSFKTLKF | HLRDVTQFHL | ARGELESLSQ | VDLASKLISM |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YGAQEAVRVV | SRSLLAMNLM | ELVDYLNQVC | LNDYREIYRE | HVRCLEERQD | WGVNSSHNKL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LLMATSSSGG | RRSPSCSDLE | QELDPVDVET | LFAPEAESYS | TPPIVVMQGS | AGTGKTTLVK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KLVQDWSKGK | LYPGQFDYVF | YVSCREVVLL | PKCDLPNLIC | WCCGDDQAPV | TEILRQPGRL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LFILDGYDEL | QKSSRAECVL | HILMRRREVP | CSLLITTRPP | ALQSLEPMLG | ERRHVLVLGF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SEEERETYFS | SCFTDKEQLK | NALEFVQNNA | VLYKACQVPG | ICWVVCSWLK | KKMARGQEVS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| ETPSNSTDIF | TAYVSTFLPT | DGNGDSSELT | RHKVLKSLCS | LAAEGMRHQR | LLFEEEVLRK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| HGLDGPSLTA | FLNCIDYRAG | LGIKKFYSFR | HISFQEFFYA | MSFLVKEDQS | QQGEATHKEV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| AKLVDPENHE | EVTLSLQFLF | DMLKTEGTLS | LGLKFCFRIA | PSVRQDLKHF | KEQIEAIKYK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RSWDLEFSLY | DSKIKKLTQG | IQMKDVILNV | QHLDEKKSDK | KKSVSVTSSF | SSGKVQSPFL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| GNDKSTRKQK | KASNGKSRGA | EEPAPGVRNR | RLASREKGHM | EMNDKEDGGV | EEQEDEEGQT |
| 670 | |||||
| LKKDGEMIDK | MNG |