Q8C0N0
Gene name |
Gm4922 |
Protein name |
Sperm motility kinase Z |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:237300 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
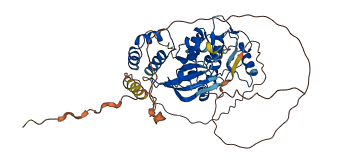
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
163-185 (Activation loop from InterPro)
Target domain |
28-275 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q8C0N0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q8C0N0-F1 | Predicted | AlphaFoldDB |
44 variants for Q8C0N0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs232250949 | 18 | M>V | No | EVA | |
| rs243766765 | 48 | H>R | No | EVA | |
| rs3389092688 | 49 | L>I | No | EVA | |
| rs3389092900 | 51 | G>D | No | EVA | |
| rs264170513 | 53 | T>P | No | EVA | |
| rs50928512 | 60 | K>N | No | EVA | |
| rs231812230 | 65 | W>C | No | EVA | |
| rs3389064421 | 84 | V>I | No | EVA | |
| rs51925484 | 93 | E>G | No | EVA | |
| rs3389078087 | 109 | H>L | No | EVA | |
| rs49611107 | 113 | Q>R | No | EVA | |
| rs3389087027 | 114 | E>A | No | EVA | |
| rs3389083752 | 181 | C>Y | No | EVA | |
| rs3389078097 | 202 | V>I | No | EVA | |
| rs3389064368 | 213 | Y>N | No | EVA | |
| rs3389092881 | 216 | T>A | No | EVA | |
| rs3389101458 | 218 | I>F | No | EVA | |
| rs3389092850 | 220 | P>Q | No | EVA | |
| rs231309130 | 232 | E>G | No | EVA | |
| rs262266858 | 232 | E>K | No | EVA | |
| rs3389068802 | 250 | S>N | No | EVA | |
| rs3389090560 | 251 | M>V | No | EVA | |
| rs3389033868 | 258 | T>A | No | EVA | |
| rs3389092698 | 267 | Q>H | No | EVA | |
| rs3389099416 | 275 | L>F | No | EVA | |
| rs3389083735 | 280 | K>N | No | EVA | |
| rs213234533 | 282 | I>V | No | EVA | |
| rs249970666 | 285 | H>Y | No | EVA | |
| rs3389099410 | 286 | S>P | No | EVA | |
| rs1131747140 | 287 | N>H | No | EVA | |
| rs1135089369 | 287 | N>I | No | EVA | |
| rs1132817836 | 288 | G>V | No | EVA | |
| rs50935046 | 289 | D>G | No | EVA | |
| rs1132499478 | 291 | S>C | No | EVA | |
| rs3389064380 | 310 | D>V | No | EVA | |
| rs3389089705 | 312 | R>I | No | EVA | |
| rs3389033882 | 317 | H>N | No | EVA | |
| rs47403792 | 339 | G>C | No | EVA | |
| rs51926525 | 352 | P>S | No | EVA | |
| rs45710395 | 357 | V>A | No | EVA | |
| rs215008568 | 406 | M>T | No | EVA | |
| rs47069570 | 408 | R>K | No | EVA | |
| rs3389092636 | 452 | S>G | No | EVA | |
| rs3389058300 | 459 | P>H | No | EVA |
No associated diseases with Q8C0N0
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
39 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P06782 | SNF1 | Carbon catabolite-derepressing protein kinase | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | EV |
| Q96L34 | MARK4 | MAP/microtubule affinity-regulating kinase 4 | Homo sapiens (Human) | SS |
| P57059 | SIK1 | Serine/threonine-protein kinase SIK1 | Homo sapiens (Human) | PR |
| Q7KZI7 | MARK2 | Serine/threonine-protein kinase MARK2 | Homo sapiens (Human) | SS |
| P27448 | MARK3 | MAP/microtubule affinity-regulating kinase 3 | Homo sapiens (Human) | SS |
| Q9P0L2 | MARK1 | Serine/threonine-protein kinase MARK1 | Homo sapiens (Human) | EV |
| O60285 | NUAK1 | NUAK family SNF1-like kinase 1 | Homo sapiens (Human) | PR |
| Q8VHJ5 | Mark1 | Serine/threonine-protein kinase MARK1 | Mus musculus (Mouse) | SS |
| Q03141 | Mark3 | MAP/microtubule affinity-regulating kinase 3 | Mus musculus (Mouse) | SS |
| Q8CIP4 | Mark4 | MAP/microtubule affinity-regulating kinase 4 | Mus musculus (Mouse) | SS |
| Q05512 | Mark2 | Serine/threonine-protein kinase MARK2 | Mus musculus (Mouse) | SS |
| Q8C0X8 | Sperm motility kinase X | Mus musculus (Mouse) | PR | |
| O35280 | Chek1 | Serine/threonine-protein kinase Chk1 | Mus musculus (Mouse) | PR |
| Q69Z98 | Brsk2 | Serine/threonine-protein kinase BRSK2 | Mus musculus (Mouse) | EV |
| Q5RJI5 | Brsk1 | Serine/threonine-protein kinase BRSK1 | Mus musculus (Mouse) | EV |
| Q61846 | Melk | Maternal embryonic leucine zipper kinase | Mus musculus (Mouse) | PR |
| O54863 | Tssk2 | Testis-specific serine/threonine-protein kinase 2 | Mus musculus (Mouse) | PR |
| Q61241 | Tssk1b | Testis-specific serine/threonine-protein kinase 1 | Mus musculus (Mouse) | PR |
| A0AUV4 | Gm7168 | Sperm motility kinase Y | Mus musculus (Mouse) | PR |
| Q641K5 | Nuak1 | NUAK family SNF1-like kinase 1 | Mus musculus (Mouse) | PR |
| Q5EG47 | Prkaa1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Mus musculus (Mouse) | SS |
| Q8BRK8 | Prkaa2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Mus musculus (Mouse) | SS |
| A2KF29 | Smoktcr | Sperm motility kinase Tcr mutant form | Mus musculus (Mouse) | PR |
| O08678 | Mark1 | Serine/threonine-protein kinase MARK1 | Rattus norvegicus (Rat) | SS |
| Q8VHF0 | Mark3 | MAP/microtubule affinity-regulating kinase 3 | Rattus norvegicus (Rat) | SS |
| O08679 | Mark2 | Serine/threonine-protein kinase MARK2 | Rattus norvegicus (Rat) | SS |
| Q8LIG4 | CIPK3 | CBL-interacting protein kinase 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q852Q1 | OSK4 | Serine/threonine protein kinase OSK4 | Oryza sativa subsp. japonica (Rice) | SS |
| Q852Q2 | OSK1 | Serine/threonine protein kinase OSK1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q6ZLP5 | CIPK23 | CBL-interacting protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAX3 | CIPK33 | CBL-interacting protein kinase 33 | Oryza sativa subsp japonica (Rice) | PR |
| Q2QY53 | CIPK32 | CBL-interacting protein kinase 32 | Oryza sativa subsp japonica (Rice) | PR |
| Q9TW45 | par-1 | Serine/threonine-protein kinase par-1 | Caenorhabditis elegans | SS |
| Q94CG0 | CIPK21 | CBL-interacting serine/threonine-protein kinase 21 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q93VD3 | CIPK23 | CBL-interacting serine/threonine-protein kinase 23 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LDI3 | CIPK24 | CBL-interacting serine/threonine-protein kinase 24 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FLZ3 | KIN12 | SNF1-related protein kinase catalytic subunit alpha KIN12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P92958 | KIN11 | SNF1-related protein kinase catalytic subunit alpha KIN11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q38997 | KIN10 | SNF1-related protein kinase catalytic subunit alpha KIN10 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MYSDSEDESS | ELSTVLSMFE | EKEFTRQYTV | LKTLSQHGTT | EVRLCSHHLT | GVTVAVKALK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YQRWWEPKVS | EVEIMKMLSH | PNIVSLLQVI | ETEQNIYLIM | EVAQGTQLHN | RVQEARCLKE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DEARSIFVQL | LSAIGYCHGE | GVVHRDLKPD | NVIVDEHGNV | KIVDFGLGAR | FMPGQKLERL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| CGAFQFIPPE | IFLGLPYDGP | KVDIWALGVL | LYYMVTGIFP | FVGSTLSEIS | KEVLQGRYEI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PYNLSKDLRS | MIGLLLATNA | RQRPTAQDLL | SHPWLQEGEK | TITFHSNGDT | SFPDPDIMAA |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MKNIGFHVQD | IRESLKHRKF | DETMATYNLL | RAEACQDDGN | YVQTKLMNPG | MPPFPSVTDS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GAFSLPPRRR | ASEPSFKVLV | SSTEEHQLRQ | TGGTNAPFPP | KKTPTMGRSQ | KQKRAMTAPC |
| 430 | 440 | 450 | 460 | 470 | 480 |
| ICLLRNTYID | TEDSSFCTSS | QAEKTSSDPE | KSETSTSCPL | TPRGWRKWKK | RIVACIQTLC |
| 490 | |||||
| CCTLPQKKCP | RSVHPQK |