Q8BTI9
Gene name |
Pik3cb |
Protein name |
Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform |
Names |
PI3-kinase subunit beta, PI3K-beta, PI3Kbeta, PtdIns-3-kinase subunit beta, Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit beta, PtdIns-3-kinase subunit p110-beta, p110beta, Serine/threonine protein kinase PIK3CB |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:74769 |
EC number |
2.7.1.153: Phosphotransferases with an alcohol group as acceptor |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
931-955 (Activation loop from InterPro)
Target domain |
700-1061 (Catalytic domain of Class IA Phosphoinositide 3-kinase beta) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

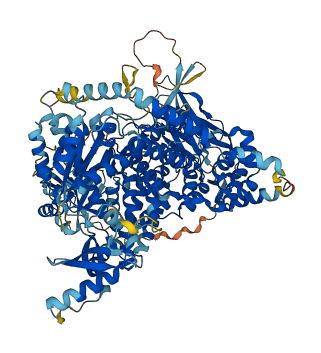
3 structures for Q8BTI9
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2Y3A | X-ray | 330 A | A | 1-1064 | PDB |
| 4BFR | X-ray | 280 A | A/B | 114-1064 | PDB |
| AF-Q8BTI9-F1 | Predicted | AlphaFoldDB |
35 variants for Q8BTI9
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389053381 | 63 | F>L | No | EVA | |
| rs3389063149 | 73 | M>I | No | EVA | |
| rs3389073421 | 76 | C>S | No | EVA | |
| rs3389010630 | 81 | A>D | No | EVA | |
| rs3389060552 | 93 | L>P | No | EVA | |
| rs3389073437 | 142 | F>V | No | EVA | |
| rs3389074247 | 153 | K>* | No | EVA | |
| rs3389066010 | 219 | E>D | No | EVA | |
| rs3389067543 | 303 | S>F | No | EVA | |
| rs249350218 | 330 | T>I | No | EVA | |
| rs3389060513 | 393 | R>* | No | EVA | |
| rs3389053410 | 403 | D>E | No | EVA | |
| rs3389067499 | 409 | K>N | No | EVA | |
| rs3389053376 | 413 | T>I | No | EVA | |
| rs3389053401 | 428 | V>A | No | EVA | |
| rs238360404 | 488 | T>K | No | EVA | |
| rs3389060596 | 569 | K>E | No | EVA | |
| rs3389040423 | 586 | Q>H | No | EVA | |
| rs3389073377 | 596 | P>A | No | EVA | |
| rs3400460507 | 599 | E>A | No | EVA | |
| rs3400226950 | 599 | E>D | No | EVA | |
| rs3411772478 | 600 | A>S | No | EVA | |
| rs3389065039 | 605 | D>H | No | EVA | |
| rs3389063123 | 635 | Q>R | No | EVA | |
| rs3389068628 | 680 | A>S | No | EVA | |
| rs33182760 | 683 | V>I | No | EVA | |
| rs3389062957 | 722 | L>P | No | EVA | |
| rs245033961 | 728 | S>N | No | EVA | |
| rs3389010608 | 738 | T>M | No | EVA | |
| rs3389060560 | 787 | R>SE* | No | EVA | |
| rs3389010573 | 788 | A>G | No | EVA | |
| rs3389060526 | 795 | G>R | No | EVA | |
| rs3389074222 | 795 | G>S | No | EVA | |
| rs3389068620 | 838 | T>R | No | EVA | |
| rs3400021817 | 865 | A>C | No | EVA |
No associated diseases with Q8BTI9
8 regional properties for Q8BTI9
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Phosphatidylinositol 3-kinase Ras-binding (PI3K RBD) domain | 174 - 282 | IPR000341 |
| domain | Phosphatidylinositol 3-/4-kinase, catalytic domain | 766 - 1061 | IPR000403 |
| domain | Phosphoinositide 3-kinase, accessory (PIK) domain | 518 - 705 | IPR001263 |
| domain | C2 phosphatidylinositol 3-kinase-type domain | 315 - 490 | IPR002420 |
| domain | Phosphatidylinositol 3-kinase, adaptor-binding domain | 20 - 112 | IPR003113 |
| conserved_site | Phosphatidylinositol 3/4-kinase, conserved site | 798 - 812 | IPR018936-1 |
| conserved_site | Phosphatidylinositol 3/4-kinase, conserved site | 898 - 918 | IPR018936-2 |
| domain | PI3Kbeta, catalytic domain | 700 - 1061 | IPR037702 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.1.153 | Phosphotransferases with an alcohol group as acceptor |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
12 GO annotations of cellular component
| Name | Definition |
|---|---|
| brush border membrane | The portion of the plasma membrane surrounding the brush border. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| phosphatidylinositol 3-kinase complex | A protein complex capable of phosphatidylinositol 3-kinase activity and containing subunits of any phosphatidylinositol 3-kinase (PI3K) enzyme. These complexes are divided in three classes (called I, II and III) that differ for their presence across taxonomic groups and for the type of their constituents. Catalytic subunits of phosphatidylinositol 3-kinase enzymes are present in all 3 classes; regulatory subunits of phosphatidylinositol 3-kinase enzymes are present in classes I and III; adaptor proteins have been observed in class II complexes and may be present in other classes too. |
| phosphatidylinositol 3-kinase complex, class IA | A class I phosphatidylinositol 3-kinase complex that possesses 1-phosphatidylinositol-4-phosphate 3-kinase activity; comprises a catalytic class IA phosphoinositide 3-kinase (PI3K) subunit and an associated SH2 domain-containing regulatory subunit that is a member of a family of related proteins often called p85 proteins. Through the interaction with the SH2-containing adaptor subunits, Class IA PI3K catalytic subunits are linked to tyrosine kinase signaling pathways. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| 1-phosphatidylinositol-3-kinase activity | Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol + ATP = a 1-phosphatidyl-1D-myo-inositol 3-phosphate + ADP + 2 H(+). |
| 1-phosphatidylinositol-4-phosphate 3-kinase activity | Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol 4-phosphate + ATP = 1-phosphatidyl-1D-myo-inositol 3,4-bisphosphate + ADP + 2 H(+). |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| insulin receptor substrate binding | Binding to an insulin receptor substrate (IRS) protein, an adaptor protein that bind to the transphosphorylated insulin and insulin-like growth factor receptors, are themselves phosphorylated and in turn recruit SH2 domain-containing signaling molecules to form a productive signaling complex. |
| kinase activity | Catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule. |
| phosphatidylinositol kinase activity | Catalysis of the reaction: ATP + a phosphatidylinositol = ADP + a phosphatidylinositol phosphate. |
| phosphatidylinositol-3,4-bisphosphate 5-kinase activity | Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol 3,4-bisphosphate + ATP = a 1-phosphatidyl-1D-myo-inositol 3,4,5-trisphosphate + ADP + 2 H(+). |
| phosphatidylinositol-4,5-bisphosphate 3-kinase activity | Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate + ATP = a 1-phosphatidyl-1D-myo-inositol 3,4,5-trisphosphate + ADP + 2 H(+). |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
28 GO annotations of biological process
| Name | Definition |
|---|---|
| angiogenesis involved in wound healing | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels and contribute to the series of events that restore integrity to a damaged tissue, following an injury. |
| autophagy | The cellular catabolic process in which cells digest parts of their own cytoplasm; allows for both recycling of macromolecular constituents under conditions of cellular stress and remodeling the intracellular structure for cell differentiation. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cellular calcium ion homeostasis | Any process involved in the maintenance of an internal steady state of calcium ions at the level of a cell. |
| embryonic cleavage | The first few specialized divisions of an activated animal egg. |
| endocytosis | A vesicle-mediated transport process in which cells take up external materials or membrane constituents by the invagination of a small region of the plasma membrane to form a new membrane-bounded vesicle. |
| endothelial cell proliferation | The multiplication or reproduction of endothelial cells, resulting in the expansion of a cell population. Endothelial cells are thin flattened cells which line the inside surfaces of body cavities, blood vessels, and lymph vessels, making up the endothelium. |
| homophilic cell adhesion via plasma membrane adhesion molecules | The attachment of a plasma membrane adhesion molecule in one cell to an identical molecule in an adjacent cell. |
| negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of hypoxia-induced intrinsic apoptotic signaling pathway. |
| negative regulation of MAP kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of MAP kinase activity. |
| negative regulation of protein kinase B signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| negative regulation of sprouting angiogenesis | Any process that stops, prevents or reduces the frequency, rate or extent of sprouting angiogenesis. |
| negative regulation of vascular endothelial growth factor signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of vascular endothelial growth factor signaling pathway. |
| phosphatidylinositol 3-kinase signaling | A series of reactions within the signal-receiving cell, mediated by the intracellular phosphatidylinositol 3-kinase (PI3K). Many cell surface receptor linked signaling pathways signal through PI3K to regulate numerous cellular functions. |
| phosphatidylinositol phosphate biosynthetic process | The chemical reactions and pathways resulting in the formation of phosphatidylinositol phosphate. |
| phosphatidylinositol-3-phosphate biosynthetic process | The chemical reactions and pathways resulting in the formation of phosphatidylinositol-3-phosphate, a phosphatidylinositol monophosphate carrying the phosphate group at the 3-position. |
| phosphatidylinositol-mediated signaling | The series of molecular signals in which a cell uses a phosphatidylinositol-mediated signaling to convert a signal into a response. Phosphatidylinositols include phosphatidylinositol (PtdIns) and its phosphorylated derivatives. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| platelet activation | A series of progressive, overlapping events triggered by exposure of the platelets to subendothelial tissue. These events include shape change, adhesiveness, aggregation, and release reactions. When carried through to completion, these events lead to the formation of a stable hemostatic plug. |
| positive regulation of endothelial cell migration | Any process that increases the rate, frequency, or extent of the orderly movement of an endothelial cell into the extracellular matrix to form an endothelium. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of neutrophil apoptotic process | Any process that activates or increases the frequency, rate, or extent of neutrophil apoptotic process. |
| positive regulation of nitric-oxide synthase activity | Any process that activates or increases the activity of the enzyme nitric-oxide synthase. |
| positive regulation of protein kinase B signaling | Any process that activates or increases the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| positive regulation of Rac protein signal transduction | Any process that activates or increases the frequency, rate or extent of Rac protein signal transduction. |
| regulation of cell-matrix adhesion | Any process that modulates the frequency, rate or extent of attachment of a cell to the extracellular matrix. |
| response to ischemia | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a inadequate blood supply. |
| sphingosine-1-phosphate receptor signaling pathway | A G protein-coupled receptor signaling pathway initiated by sphingosine-1-phosphate binding to its receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P32871 | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | Bos taurus (Bovine) | PR |
| P42336 | PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | Homo sapiens (Human) | PR |
| P48736 | PIK3CG | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | Homo sapiens (Human) | PR |
| O00329 | PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | Homo sapiens (Human) | PR |
| P42338 | PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | Homo sapiens (Human) | PR |
| P42337 | Pik3ca | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | Mus musculus (Mouse) | PR |
| Q9JHG7 | Pik3cg | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | Mus musculus (Mouse) | PR |
| Q8BKC8 | Pi4kb | Phosphatidylinositol 4-kinase beta | Mus musculus (Mouse) | PR |
| Q61194 | Pik3c2a | Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | Mus musculus (Mouse) | PR |
| O35904 | Pik3cd | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | Mus musculus (Mouse) | PR |
| O02697 | PIK3CG | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | Sus scrofa (Pig) | PR |
| Q9Z1L0 | Pik3cb | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | Rattus norvegicus (Rat) | PR |
| Q94125 | age-1 | Phosphatidylinositol 3-kinase age-1 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPPAMADNLD | IWAVDSQIAS | DGAISVDFLL | PTGIYIQLEV | PREATISYIK | QMLWKQVHNY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PMFNLLMDID | SYMFACVNQT | AVYEELEDET | RRLCDVRPFL | PVLKLVTRSC | DPAEKLDSKI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GVLIGKGLHE | FDALKDPEVN | EFRRKMRKFS | EAKIQSLVGL | SWIDWLKHTY | PPEHEPSVLE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NLEDKLYGGK | LVVAVHFENS | QDVFSFQVSP | NLNPIKINEL | AIQKRLTIRG | KEDEASPCDY |
| 250 | 260 | 270 | 280 | 290 | 300 |
| VLQVSGRVEY | VFGDHPLIQF | QYIRNCVMNR | TLPHFILVEC | CKIKKMYEQE | MIAIEAAINR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NSSNLPLPLP | PKKTRVISHI | WDNNNPFQIT | LVKGNKLNTE | ETVKVHVRAG | LFHGTELLCK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TVVSSEISGK | NDHIWNEQLE | FDINICDLPR | MARLCFAVYA | VLDKVKTKKS | TKTINPSKYQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TIRKAGKVHY | PVAWVNTMVF | DFKGQLRSGD | VILHSWSSFP | DELEEMLNPM | GTVQTNPYAE |
| 490 | 500 | 510 | 520 | 530 | 540 |
| NATALHITFP | ENKKQPCYYP | PFDKIIEKAA | ELASGDSANV | SSRGGKKFLA | VLKEILDRDP |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LSQLCENEMD | LIWTLRQDCR | ENFPQSLPKL | LLSIKWNKLE | DVAQLQALLQ | IWPKLPPREA |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LELLDFNYPD | QYVREYAVGC | LRQMSDEELS | QYLLQLVQVL | KYEPFLDCAL | SRFLLERALD |
| 670 | 680 | 690 | 700 | 710 | 720 |
| NRRIGQFLFW | HLRSEVHTPA | VSVQFGVILE | AYCRGSVGHM | KVLSKQVEAL | NKLKTLNSLI |
| 730 | 740 | 750 | 760 | 770 | 780 |
| KLNAVKLSRA | KGKEAMHTCL | KQSAYREALS | DLQSPLNPCV | ILSELYVEKC | KYMDSKMKPL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| WLVYSSRAFG | EDSVGVIFKN | GDDLRQDMLT | LQMLRLMDLL | WKEAGLDLRM | LPYGCLATGD |
| 850 | 860 | 870 | 880 | 890 | 900 |
| RSGLIEVVST | SETIADIQLN | SSNVAATAAF | NKDALLNWLK | EYNSGDDLDR | AIEEFTLSCA |
| 910 | 920 | 930 | 940 | 950 | 960 |
| GYCVASYVLG | IGDRHSDNIM | VKKTGQLFHI | DFGHILGNFK | SKFGIKRERV | PFILTYDFIH |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| VIQQGKTGNT | EKFGRFRQCC | EDAYLILRRH | GNLFITLFAL | MLTAGLPELT | SVKDIQYLKD |
| 1030 | 1040 | 1050 | 1060 | ||
| SLALGKSEEE | ALKQFKQKFD | EALRESWTTK | VNWMAHTVRK | DYRS |